| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,096,647 – 11,096,752 |
| Length | 105 |
| Max. P | 0.980230 |

| Location | 11,096,647 – 11,096,752 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 64.83 |
| Shannon entropy | 0.76608 |
| G+C content | 0.38090 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -12.14 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11096647 105 - 23011544 CAAGUCCAUCUAUAAAUGCUUAUA----AUUAAGCGUUUUUUU---UUCAUUACUUAGCAAUUUCUGGAUACUUUUGGCAAGUCCGUGUUGAUUGGUGUUCAAACCAGCCUA .((((........(((((((((..----..)))))))))....---......)))).((......((((((((.(..(((......)))..)..)))))))).....))... ( -20.57, z-score = -1.27, R) >droSim1.chr2L 10895683 105 - 22036055 CAAGUCCAUCUAUAGAUGCUUAUA----AUUCGAAUUUUAACU---CAUUUCUAUUAGCAAUUUCUGGACACUUUUGGCAAGUCCGUGUUGAUUGGUGUUCAAACCAGCCUC ...(((((((....))((((.(((----....(((........---...)))))).)))).....)))))......(((.((((......))))(((......))).))).. ( -17.60, z-score = -0.04, R) >droSec1.super_3 6497976 104 - 7220098 CAAGUCCAUCCA-AGAUGCUUAUA----AUUCGAAUUUUAACU---CAUUUCUAUUAGCAAUUUCUGGACACUUUUGGCAAGUCCGUGUUGAUUGGUGUUCAAACCAGCCUU ...(((((...(-((.((((.(((----....(((........---...)))))).)))).))).)))))......(((.((((......))))(((......))).))).. ( -18.20, z-score = -0.18, R) >droYak2.chr2L 7492627 105 - 22324452 UUAGUCCACCUAUAGAUUCUUAUU----AUUAAGAGUUUAAUU---CAUUUCCGGUAGCAAUUUCUGGACACUUUUGGCAAGUCCGUGCUGAUUGGUGUCCAAACCAGCCUG ............((((((((((..----..))))))))))...---.......(((.........((((((((.(..(((......)))..)..)))))))).....))).. ( -26.64, z-score = -2.45, R) >droEre2.scaffold_4929 12293117 105 + 26641161 CGGGUCCAUCUAUAGAUGCUUAUA----ACUAAGAGUUUAAUU---CAUUUCCACUAGCAAUUUCUGGACACUUUUGGCAAGUCCGUGUUGAUUGGUGUCCAAACCAGCCUU .((((...........((((....----.....((((...)))---).........)))).....((((((((.(..(((......)))..)..)))))))).....)))). ( -22.87, z-score = -0.52, R) >dp4.chr4_group3 6445979 108 - 11692001 CAAGUAAAGCUCCUAAUUGUCCACUAUUGAUAU-UAUUUUGUG---AUACUCUACUAGCAAUUUAUUGAUUCCUUCGGCAAGUCAGUGCUGAUAGGUGUCCAAACCAGCCUA ........(((......((((.......)))).-...((((.(---((((.(((.(((((.....((((((.(....)..))))))))))).))))))))))))..)))... ( -20.50, z-score = -0.45, R) >droPer1.super_1 7937499 108 - 10282868 CAAGUAAAGCUCCUAAUUGUCCACUAUUGAUAU-UAUUUUGUG---AUACUCUACUAGCAAUUUAUUGAUUCCUUCGGCAAGUCAGUGCUGAUAGGUGUCCAAACCAGCCUA ........(((......((((.......)))).-...((((.(---((((.(((.(((((.....((((((.(....)..))))))))))).))))))))))))..)))... ( -20.50, z-score = -0.45, R) >droWil1.scaffold_181132 406779 105 + 1035393 ----CUGAAACAAAAAUAAUGAAUUCCUAAUAUAAUUUAUGUA---CAUUUUUUGUAGCAAUUUUUGGACUCAUUUGGAAAAUCCGUAUUGCUUGGCGUCCAAGCCAGCCUG ----.....(((((((..(((((((........)))))))...---...)))))))((((((...((((.((.....))...)))).))))))((((......))))..... ( -19.20, z-score = -0.89, R) >droVir3.scaffold_12963 12267721 104 + 20206255 AAAAUUCAAUCGAA--UUGUCGUGUCGUAACAACAACUAUUUG---CAUUG---ACAGCAAUUUCUGGAUGCCUUUGGCAAAUCUGUGCUGAUUGGCGUUCAGACAAGCCUG .....(((((((((--(.((..(((....)))...)).)))))---.))))---)(((.....((((((((((.(..(((......)))..)..)))))))))).....))) ( -29.70, z-score = -2.18, R) >droMoj3.scaffold_6500 2920589 107 - 32352404 --AAUAUAAACGGAGUCUCUUCUAUUGCAAUAACAAUUUUCUA---AUUUGUGUUCAGCAAUUCCUGGAUGCCUUUGGCAAAUCCGUGCUGAUUGGCGUGCAGACAAGUCUG --........((((.........(((((....((((.......---..)))).....)))))..(((.(((((.(..(((......)))..)..))))).))).....)))) ( -25.00, z-score = -0.95, R) >droGri2.scaffold_15252 6185651 107 + 17193109 -----AAAACCCCAAAUUUUCCGCUUUAUAUAUAUAAUUUACAUAUUUGUAUUUCUAGCAAUUCCUGGACGCUUUUGGCAAGUCCGUGCUGAUUGGCGUACAGACAAGCCUG -----.................(((..(((((.(((.......))).)))))....))).....(((.(((((.(..(((......)))..)..))))).)))......... ( -20.40, z-score = -0.88, R) >consensus CAAGUCCAACUAUAAAUUCUUAUAU___AAUAAAAAUUUUAUU___CAUUUCUACUAGCAAUUUCUGGACACUUUUGGCAAGUCCGUGCUGAUUGGUGUCCAAACCAGCCUG .........................................................((...((.((((((((.((((((......))))))..))))))))))...))... (-12.14 = -11.28 + -0.85)

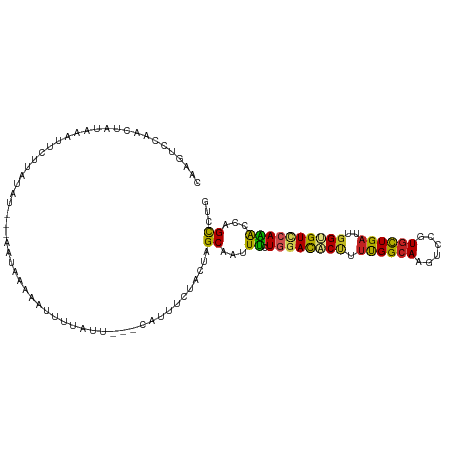
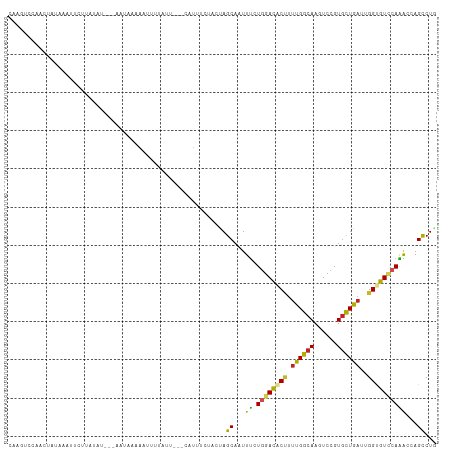
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:32 2011