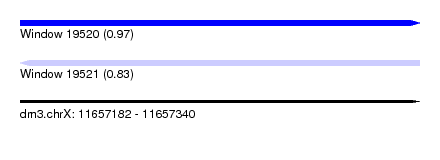
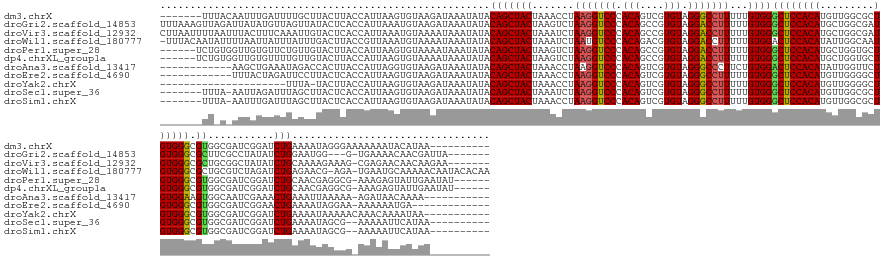
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,657,182 – 11,657,340 |
| Length | 158 |
| Max. P | 0.973976 |

| Location | 11,657,182 – 11,657,340 |
|---|---|
| Length | 158 |
| Sequences | 11 |
| Columns | 175 |
| Reading direction | forward |
| Mean pairwise identity | 73.93 |
| Shannon entropy | 0.54785 |
| G+C content | 0.40920 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -25.29 |
| Energy contribution | -24.66 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
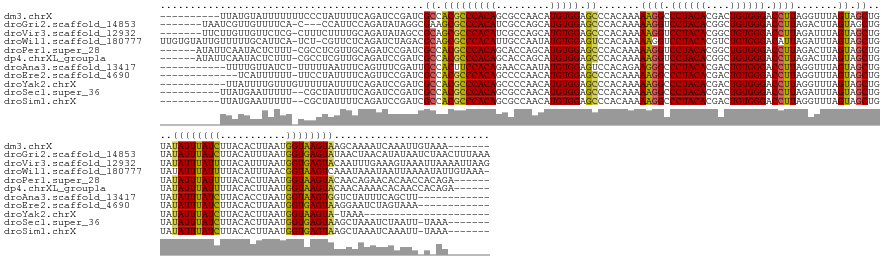
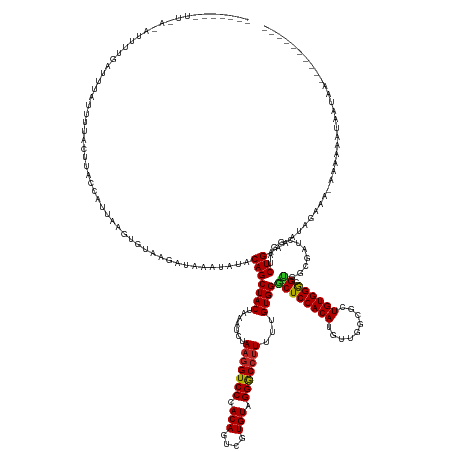
>dm3.chrX 11657182 158 + 22422827 ----------UUAUGUAUUUUUUUCCCUAUUUUCAGAUCCGAUCGCCACGCCCACAGCGCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUAAGUAAGCAAAAUCAAAUUGUAAA------- ----------..............................(((.((.((.(((((((((......(((((.(((........))).))))))).)))))))(((...)))...)).))(((.((((((((...........)))))))).)))..)))..........------- ( -33.70, z-score = -0.35, R) >droGri2.scaffold_14853 8339836 164 - 10151454 -------UAAUCGUUGUUUUCA-C---CCAUUCCAGAUAUAGGCGAAGCGCCCACAUCGCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUCUUACAUUUAAUGGUGAGUAUAACUAACAUAUAAUCUAACUUUAAA -------.....(((((.((((-(---(......(((((..(((((.(......).)))))(((.(((((...)))))..(((((((((((....)))))))))))..........))).......)))))..........)))))).)))))...................... ( -45.39, z-score = -2.76, R) >droVir3.scaffold_12932 1462053 167 - 2102469 -------UUCUUGUUGUUCUCG-CUUUCUUUUGCAGAUAUAGCCGCAGCGCCCACAUCGCCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUUUUACAUUUAAUGGUGAGUACAAUUUGAAAGUAAAUUAAAAUUAAG -------(((..(((((.((((-((......((.(((((.....((((((((((((((....).)))))).)).......(((((((((((....)))))))))))..........))))).....)))))..))......)))))).)))))..)))................. ( -50.60, z-score = -3.51, R) >droWil1.scaffold_180777 378183 172 + 4753960 UUGUGUAUUGUUUUUGCAUUCA-UCU-CGUUCUCAGAUCUAGACGCAGCGCCCACAUUGCCAAUAUGUGGAGUCCACAAAAAGGUCCUACACGUCUGUGGGACAUUAGAUUUAGUAGCUGUAUAUUUAUUUUACAUUUAACGGUAAGUCAAAUAAAUAAUUAAAAUAUUGUAAA- ............((((((....-...-.(((((.((((((((.....(.((((((((.......)))))).)).)........((((((((....)))))))).)))))))))).)))(((.(((((..(((((........)))))..))))).)))..........))))))- ( -35.50, z-score = -0.62, R) >droPer1.super_28 409192 162 - 1111753 ------AUAUUCAAUACUCUUU-CGCCUCGUUGCAGAUCCGAUCGCCACGCCCACAGCACCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUUUUACACUUAAUGGUAAGUACAACAGAACACAACCACAGA------ ------...(((.((((....(-((..((......))..)))..((.(((((((((((....)).))))).)).......(((((((((((....))))))))))).......)).)).))))...........(((((....)))))......)))............------ ( -38.30, z-score = -0.99, R) >dp4.chrXL_group1a 6885011 162 - 9151740 ------AUAUUCAAUACUCUUU-CGCCUCGUUGCAGAUCCGAUCGCCACGCCCACAGCACCAGCAUGUGGAGCCCACAAAAAGGUCCUACACGGCUGUGGGACCUUAGACUUAGUAGCUGUAUAUUUAUUUUACACUUAAUGGUAAGUACAACAAAACACAACCACAGA------ ------.......((((....(-((..((......))..)))..((.(((((((((((....)).))))).)).......(((((((((((....))))))))))).......)).)).)))).................((((..((........))...))))....------ ( -37.70, z-score = -1.07, R) >droAna3.scaffold_13417 1404370 151 + 6960332 -----------UUUUGUUAUCU-UUUUUAAUUUCAGUUUCGAUUGCCACUUCCACAGAACCAAUAUGUGGAGUCCACAGAAGGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACCUAAUGGUAAGUGGUCUAUUUCAGCUU------------ -----------...........-...........((((..(((.((((((((((..(((((....(((((...))))).((((..((((((....)))))).)))).)))))..(((.((((.........)))).))).))).)))))))..)))..)))).------------ ( -39.50, z-score = -1.68, R) >droEre2.scaffold_4690 15206482 149 - 18748788 -------------UCAUUUUUU-UUCCUAUUUUCAGUUCCGAUCGCCACGCCCACAGCCCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUGAGUAAGGAAUCUAGUAAA------------ -------------.........-(((((.....(((((.(....(((..(((((((.........))))).)).......((((.((((((....)))))).)))).)))...).)))))..((((((((...........))))))))))))).........------------ ( -37.20, z-score = -1.29, R) >droYak2.chrX 6729769 142 - 21770863 -----------UUAUUUUGUUUGUUUUUAUUUUCAGAUCCGAUCGCCACGCCCACAGCCCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUAAGUA-UAAA--------------------- -----------......................(((........(((..(((((((.........))))).)).......((((.((((((....)))))).)))).))).......)))((((((((((...........))))))))-))..--------------------- ( -31.76, z-score = -0.54, R) >droSec1.super_36 332894 155 + 449511 ----------UUAUGAAUUUUU--CGCUAUUUUCAGAUCCGAUCGCCACGCCCACAGCGCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUGAGUAAGCUAAAUCUAAUU-UAAA------- ----------...((((..(..--....)..))))...............(((((((((......(((((.(((........))).))))))).)))))))...(((((((((((.((((((....)))....((((....)))))))..)))))))))))..-....------- ( -37.10, z-score = -1.71, R) >droSim1.chrX 8973946 155 + 17042790 ----------UUAUGAAUUUUU--CGCUAUUUUCAGAUCCGAUCGCCACGCCCACAGCGCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGGUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUGAGUAAGCUAAAUCAAAUU-UAAA------- ----------...(((((((..--.((((((.........((((......(((((((((......(((((.(((........))).))))))).)))))))......)))).)))))).((.((((((((...........)))))))).))......)))))-))..------- ( -37.10, z-score = -1.22, R) >consensus __________UUAUUAUUUUUU_UCCCUAUUUUCAGAUCCGAUCGCCACGCCCACAGCGCCAACAUGUGGAGCCCACAAAAAGGCCCUACACGACUGUGGGACCUUAGAUUUAGUAGCUGUAUAUUUAUCUUACACUUAAUGGUAAGUAAAAUAAAUCAAAAA_U_AA_______ ............................................((.(((((((((.........))))).)).......((((.((((((....)))))).)))).......)).))....((((((((...........)))))))).......................... (-25.29 = -24.66 + -0.62)
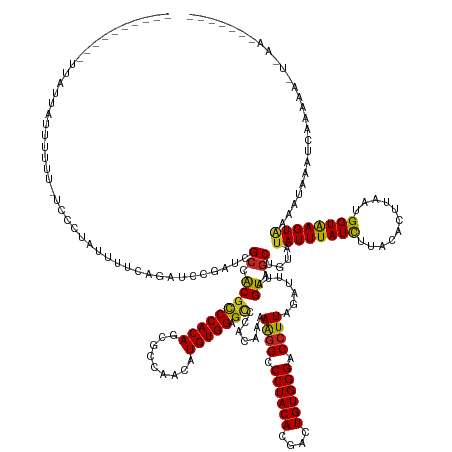
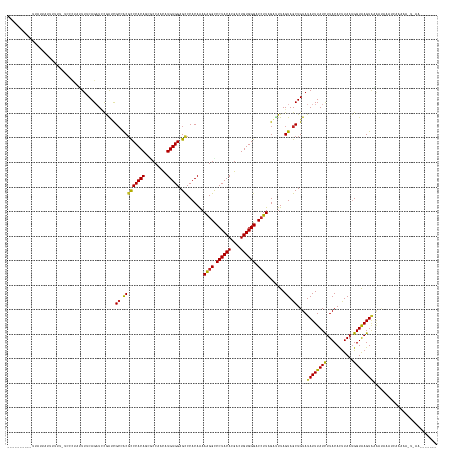
| Location | 11,657,182 – 11,657,340 |
|---|---|
| Length | 158 |
| Sequences | 11 |
| Columns | 175 |
| Reading direction | reverse |
| Mean pairwise identity | 73.93 |
| Shannon entropy | 0.54785 |
| G+C content | 0.40920 |
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.97 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11657182 158 - 22422827 -------UUUACAAUUUGAUUUUGCUUACUUACCAUUAAGUGUAAGAUAAAUAUACAGCUACUAAACCUAAGGUCCCACAGUCGUGUAGGGCCUUUUUGUGGGCUCCACAUGUUGGCGCUGUGGGCGUGGCGAUCGGAUCUGAAAAUAGGGAAAAAAAUACAUAA---------- -------....((((((((((.......((((((.....).)))))...........(((((...(((...)))(((((((((((((.((((((......)))))).))))).....)))))))).))))))))))))).)).......................---------- ( -43.30, z-score = -1.25, R) >droGri2.scaffold_14853 8339836 164 + 10151454 UUUAAAGUUAGAUUAUAUGUUAGUUAUACUCACCAUUAAAUGUAAGAUAAAUAUACAGCUACUAAGUCUAAGGUCCCACAGCCGUGUAGGACCUUUUUGUGGGCUCCACAUGCUGGCGAUGUGGGCGCUUCGCCUAUAUCUGGAAUGG---G-UGAAAACAACGAUUA------- .................((((..((.(((((((((....................((((..........(((((((.(((....))).)))))))..(((((...))))).))))..((((((((((...)))))))))))))..)))---)-)).))..))))....------- ( -45.40, z-score = -1.38, R) >droVir3.scaffold_12932 1462053 167 + 2102469 CUUAAUUUUAAUUUACUUUCAAAUUGUACUCACCAUUAAAUGUAAAAUAAAUAUACAGCUACUAAAUCUAAGGUCCCACAGCCGUGUAGGACCUUUUUGUGGGCUCCACAUGCUGGCGAUGUGGGCGCUGCGGCUAUAUCUGCAAAAGAAAG-CGAGAACAACAAGAA------- .......................((((.(((........................((((..(((.....(((((((.(((....))).)))))))....)))((.(((((((....).))))))))))))..(((...(((.....))).))-)))).))))......------- ( -40.00, z-score = -0.90, R) >droWil1.scaffold_180777 378183 172 - 4753960 -UUUACAAUAUUUUAAUUAUUUAUUUGACUUACCGUUAAAUGUAAAAUAAAUAUACAGCUACUAAAUCUAAUGUCCCACAGACGUGUAGGACCUUUUUGUGGACUCCACAUAUUGGCAAUGUGGGCGCUGCGUCUAGAUCUGAGAACG-AGA-UGAAUGCAAAAACAAUACACAA -......((((((........((((((((.....))))))))......))))))...((......((((...((((.(((....))).))))(((..(.(((((.((((((.......))))))((...))))))).)...)))....-)))-)....))............... ( -30.84, z-score = 0.21, R) >droPer1.super_28 409192 162 + 1111753 ------UCUGUGGUUGUGUUCUGUUGUACUUACCAUUAAGUGUAAAAUAAAUAUACAGCUACUAAGUCUAAGGUCCCACAGCCGUGUAGGACCUUUUUGUGGGCUCCACAUGCUGGUGCUGUGGGCGUGGCGAUCGGAUCUGCAACGAGGCG-AAAGAGUAUUGAAUAU------ ------(((((((((((((..((((.((((((....))))))...))))...))))))))))...((((.....(((((((((((((.((((((......))).)))))))).....))))))))(((.(((........))).))))))).-..)))...........------ ( -50.20, z-score = -1.13, R) >dp4.chrXL_group1a 6885011 162 + 9151740 ------UCUGUGGUUGUGUUUUGUUGUACUUACCAUUAAGUGUAAAAUAAAUAUACAGCUACUAAGUCUAAGGUCCCACAGCCGUGUAGGACCUUUUUGUGGGCUCCACAUGCUGGUGCUGUGGGCGUGGCGAUCGGAUCUGCAACGAGGCG-AAAGAGUAUUGAAUAU------ ------((.(((.((.((((((((((((..(.((.....(((((.......))))).(((((..((((((((((((.(((....))).)))))).....))))))(((((.((....)))))))..)))))....)))..))))))))))))-...)).))).))....------ ( -52.20, z-score = -1.78, R) >droAna3.scaffold_13417 1404370 151 - 6960332 ------------AAGCUGAAAUAGACCACUUACCAUUAGGUGUAAGAUAAAUAUACAGCUACUAAACCUAAGGUCCCACAGUCGUGUAGGGCCCUUCUGUGGACUCCACAUAUUGGUUCUGUGGAAGUGGCAAUCGAAACUGAAAUUAAAAA-AGAUAACAAAA----------- ------------...........(.((((((.(((((((.((((.........)))).)))...((((.((((.((((((....))).))).)))).(((((...)))))....))))..))))))))))).....................-...........----------- ( -33.40, z-score = -0.32, R) >droEre2.scaffold_4690 15206482 149 + 18748788 ------------UUUACUAGAUUCCUUACUCACCAUUAAGUGUAAGAUAAAUAUACAGCUACUAAACCUAAGGUCCCACAGUCGUGUAGGGCCUUUUUGUGGGCUCCACAUGUUGGGGCUGUGGGCGUGGCGAUCGGAACUGAAAAUAGGAA-AAAAAAUGA------------- ------------.........(((((...(((((.....(((((.......))))).(((((....((((.(((((((....(((((.((((((......)))))).))))).))))))).)))).)))))....))...)))....)))))-.........------------- ( -46.90, z-score = -2.30, R) >droYak2.chrX 6729769 142 + 21770863 ---------------------UUUA-UACUUACCAUUAAGUGUAAGAUAAAUAUACAGCUACUAAACCUAAGGUCCCACAGUCGUGUAGGGCCUUUUUGUGGGCUCCACAUGUUGGGGCUGUGGGCGUGGCGAUCGGAUCUGAAAAUAAAAACAAACAAAAUAA----------- ---------------------((((-((((((....))))))))))...........(((((....((((.(((((((....(((((.((((((......)))))).))))).))))))).)))).))))).................................----------- ( -43.90, z-score = -2.72, R) >droSec1.super_36 332894 155 - 449511 -------UUUA-AAUUAGAUUUAGCUUACUCACCAUUAAGUGUAAGAUAAAUAUACAGCUACUAAAUCUAAGGUCCCACAGUCGUGUAGGGCCUUUUUGUGGGCUCCACAUGUUGGCGCUGUGGGCGUGGCGAUCGGAUCUGAAAAUAGCG--AAAAAUUCAUAA---------- -------....-..((((((((..(((((.(........).)))))...........(((((((....))..(.(((((((((((((.((((((......)))))).))))).....))))))))))))))....))))))))........--............---------- ( -42.50, z-score = -1.44, R) >droSim1.chrX 8973946 155 - 17042790 -------UUUA-AAUUUGAUUUAGCUUACUCACCAUUAAGUGUAAGAUAAAUAUACAGCUACUAAACCUAAGGUCCCACAGUCGUGUAGGGCCUUUUUGUGGGCUCCACAUGUUGGCGCUGUGGGCGUGGCGAUCGGAUCUGAAAAUAGCG--AAAAAUUCAUAA---------- -------((((-.((((((((...(((((.(........).)))))...........(((((...(((...)))(((((((((((((.((((((......)))))).))))).....)))))))).))))))))))))).)))).......--............---------- ( -44.80, z-score = -1.86, R) >consensus _______UU_A_AUUUUGAUUUAUUUUACUUACCAUUAAGUGUAAGAUAAAUAUACAGCUACUAAAUCUAAGGUCCCACAGUCGUGUAGGGCCUUUUUGUGGGCUCCACAUGUUGGCGCUGUGGGCGUGGCGAUCGGAUCUGAAAAUAGAAA_AAAAAAUAAUAA__________ .......................................................(((((((.......(((((((.(((....))).)))))))...))))((((((((.........)))))).))...........)))................................. (-22.24 = -21.97 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:21 2011