
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,637,003 – 11,637,099 |
| Length | 96 |
| Max. P | 0.781847 |

| Location | 11,637,003 – 11,637,099 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.12 |
| Shannon entropy | 0.34956 |
| G+C content | 0.41309 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.32 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11637003 96 + 22422827 ---GUGAGUGGUAGCGGUGC-AAAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC--------- ---..(((((((((((((((-((((((.((..(......)..))..)))))...))))))))).)))))))..............((....)).......--------- ( -30.30, z-score = -2.74, R) >droSim1.chrX 8966273 96 + 17042790 ---GUGAGUGGUAGCGGUGC-AAAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC--------- ---..(((((((((((((((-((((((.((..(......)..))..)))))...))))))))).)))))))..............((....)).......--------- ( -30.30, z-score = -2.74, R) >droSec1.super_36 325687 97 + 449511 ---GUGAGUGGUAGCGGUGC-AAAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUCC-------- ---..(((((((((((((((-((((((.((..(......)..))..)))))...))))))))).)))))))..............((....))........-------- ( -30.30, z-score = -2.73, R) >droYak2.chrX 6722884 96 - 21770863 ---GUGAGUGGUAGCGGUGC-AAAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC--------- ---..(((((((((((((((-((((((.((..(......)..))..)))))...))))))))).)))))))..............((....)).......--------- ( -30.30, z-score = -2.74, R) >droEre2.scaffold_4690 15198869 96 - 18748788 ---GUGAGUGGUAGCGGUGC-AAAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC--------- ---..(((((((((((((((-((((((.((..(......)..))..)))))...))))))))).)))))))..............((....)).......--------- ( -30.30, z-score = -2.74, R) >droAna3.scaffold_13417 1394644 99 + 6960332 GUGAGUGGUAAGAGCGGUGC-UAAUGCAUCUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCCAAUGCGACAAUC--------- ..(((((((...((((((((-((((((.((..(......)..))..))))))...)))))))).)))))))..............((....)).......--------- ( -30.00, z-score = -2.55, R) >dp4.chrXL_group1a 6877409 96 - 9151740 ---GUGAGUGGUAGCGGUGU-UGAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC--------- ---..(((((((((((((((-((((((.((..(......)..))..))))))...)))))))).)))))))..............((....)).......--------- ( -28.70, z-score = -2.19, R) >droPer1.super_6843 387 96 + 1592 ---GUGAGUGGUAGCGGUGU-UGAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC--------- ---..(((((((((((((((-((((((.((..(......)..))..))))))...)))))))).)))))))..............((....)).......--------- ( -28.70, z-score = -2.19, R) >droWil1.scaffold_180777 369925 99 + 4753960 ---GUGAGUCUUAAUGGAGCUUAAUACGUU---UUCGAUGUAAGAUGAUUUAAAUGCACCGUUAACUACAUUUCAUUCUAAUUAAGUUAAUGAAUCGAUAGAAAC---- ---.......(((((((.((((((..((((---((......))))))..))))..)).)))))))((((..((((((((.....))..))))))..).)))....---- ( -13.50, z-score = 0.68, R) >droVir3.scaffold_12932 1453817 91 - 2102469 ---------ACGAUCGGUGUUAAAUGCAUCUGAUCCGUCUUGGAGCGCAUUAAAUACACCGUUAACUACUUCGAAUUCUAAUCAAGCUGAUGCGACAAAC--------- ---------.(((.((((((..(((((.((..(......)..))..)))))....)))))).........)))............((....)).......--------- ( -18.70, z-score = -0.84, R) >droMoj3.scaffold_6473 894651 91 + 16943266 ---------AAGAUCGGUGUUAAAUGCAUUUGAUCCGUCUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUCAAGCUGAUGCGACAAAC--------- ---------.(((((((.(((((.((((((((((.(((......))).))))))))))...)))))....))))..)))......((....)).......--------- ( -21.40, z-score = -1.39, R) >droGri2.scaffold_14853 8327948 100 - 10151454 ---------GAGAUCGGUGUUAAAUGCAUUUGAUCCGUCUUGGAGCGCAUUAAAUGCACCGUUAACUACCUCGAAUUCUAAUCAAGCUGAUGCGACAAAAAGAAAAAGA ---------......((((((((.((((((((((.(((......))).))))))))))...))))).))).....((((......((....)).......))))..... ( -23.02, z-score = -1.36, R) >consensus ___GUGAGUGGUAGCGGUGC_AAAUGCAUUUGAUCCGUUUUGGAGCGCAUUAAAUGCACCGUUAACUACUUCGAAUUCUAAUUAAGCUGAUGCGACAAUC_________ ..............((((((..(((((.((..(......)..))..)))))....))))))........................((....))................ (-15.80 = -15.32 + -0.48)
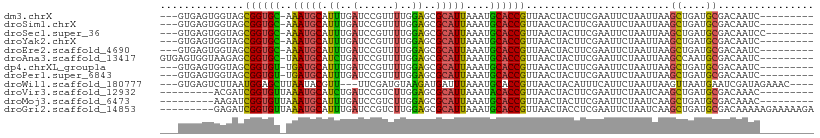

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:18 2011