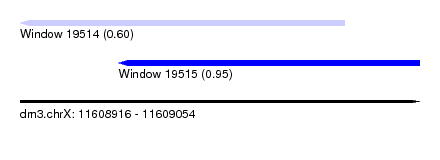
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,608,916 – 11,609,054 |
| Length | 138 |
| Max. P | 0.953701 |

| Location | 11,608,916 – 11,609,028 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.02 |
| Shannon entropy | 0.13433 |
| G+C content | 0.51007 |
| Mean single sequence MFE | -10.80 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
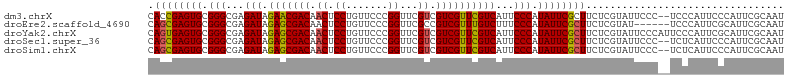
>dm3.chrX 11608916 112 - 22422827 CAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC--UCCCAUUCCCAUUCGCAAUCCCAAUUCCCAAUUCCCGUCACACGAGUUAGCAG .(((((.(((...(((...(.((.....)).)............((.............--..............))..................)))..))).)))))..... ( -10.53, z-score = -1.96, R) >droEre2.scaffold_4690 15170606 108 + 18748788 CAACUCCUGUUCCCGGUUCGCCGUCGUUUGUCUUUCCCAUAUUCGCUUCUCGUAUUCCCAUUCGCAUUCGCAAUCCCAA------UUCCCAAUUCCCGUCACACGAGUUAGCAG ......(((((..(((....)).((((.((.(............((.....))..........((....))........------............).)).)))))..))))) ( -11.80, z-score = -1.24, R) >droYak2.chrX 6693850 114 + 21770863 CAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCCAUUCCCAUUCGCAUUCGCAAUCCCAAUUCCCAAUUCUCGUCACACGAGUUAGCAG ......(((((...((...(.((.....)).).....................................((....))....))...........((((.....))))..))))) ( -10.60, z-score = -1.05, R) >droSec1.super_36 297953 105 - 449511 CAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC--UCUCAUUCCCAUUCGCAA-------UCCCAAUUCCCGUCACACGAGUUAGCAG .(((((.(((...(((...(.((.....)).)............((.............--..............))..-------.........)))..))).)))))..... ( -10.53, z-score = -1.59, R) >droSim1.chrX 8938808 112 - 17042790 CAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC--UCUCAUUCCCAUUCGCAAUCCCAAUUCCCAAUUCCCGUCACACGAGUUAGCAG .(((((.(((...(((...(.((.....)).)............((.............--..............))..................)))..))).)))))..... ( -10.53, z-score = -1.84, R) >consensus CAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC__UCCCAUUCCCAUUCGCAAUCCCAAUUCCCAAUUCCCGUCACACGAGUUAGCAG .(((((.(((...(((...(.((.....)).)............((.....))..........................................)))..))).)))))..... ( -9.32 = -9.00 + -0.32)

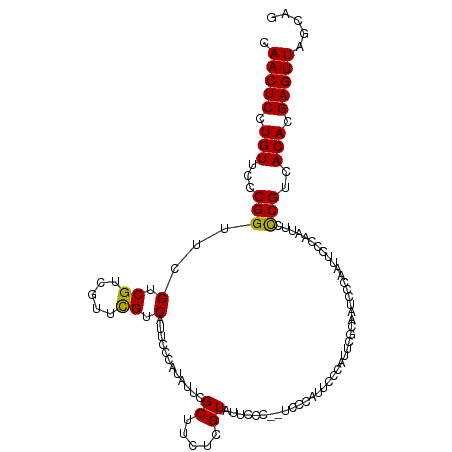
| Location | 11,608,950 – 11,609,054 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Shannon entropy | 0.10005 |
| G+C content | 0.53674 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11608950 104 - 22422827 CACCGAGUGCGGGCGAGAUAGAACGACAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC--UCCCAUUCCCAUUCGCAAU ....(((((((((((((((.(((((((.((.((.......))...)).))))))))))..........)))))...)))))))..--................... ( -24.40, z-score = -1.30, R) >droEre2.scaffold_4690 15170640 100 + 18748788 CAGCGAGUGCGGGCGAGAUAGAGCGACAACUCCUGUUCCCGGUUCGCCGUCGUUUGUCUUUCCCAUAUUCGCUUCUCGUAU------UCCCAUUCGCAUUCGCAAU ..(((((((((((.((((((((.(((((((....)))...((....)))))))))))))).)))......((.....))..------........))))))))... ( -35.50, z-score = -3.74, R) >droYak2.chrX 6693884 106 + 21770863 CAGUGAGUGCGGGCGAGAUAGAGCGACAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCCAUUCCCAUUCGCAUUCGCAAU ..((((((((((((((((..(((((((.((.((.......))...)).)))))))((.............)).))))))..............))))))))))... ( -30.45, z-score = -2.49, R) >droSec1.super_36 297980 104 - 449511 CAGCGAGUGCGGGCGAGAUAGAGCGACAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC--UCUCAUUCCCAUUCGCAAU ..(((((((.(((.((((..(((((((.((.((.......))...)).)))))))...............((.....))......--))))..))))))))))... ( -29.10, z-score = -2.21, R) >droSim1.chrX 8938842 104 - 17042790 CAGCGAGUGCGGGCGAGAUAGAGCGACAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC--UCUCAUUCCCAUUCGCAAU ..(((((((.(((.((((..(((((((.((.((.......))...)).)))))))...............((.....))......--))))..))))))))))... ( -29.10, z-score = -2.21, R) >consensus CAGCGAGUGCGGGCGAGAUAGAGCGACAACUCCUGUUCCCGGUUCGUCGUCGUUCGUCAUUCCCAUAUUCGCUUCUCGUAUUCCC__UCCCAUUCCCAUUCGCAAU .((((((((.(((...(((.(((((((.((.((.......))...)).))))))))))...))).))))))))................................. (-26.10 = -26.02 + -0.08)
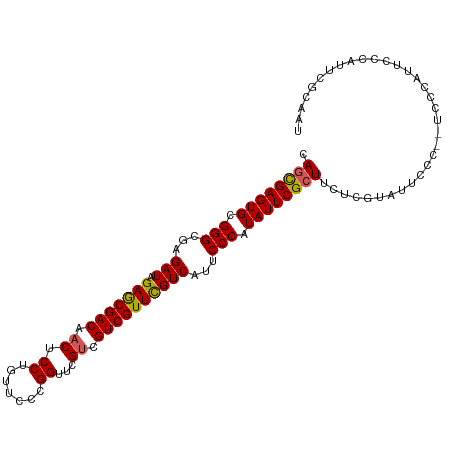
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:15 2011