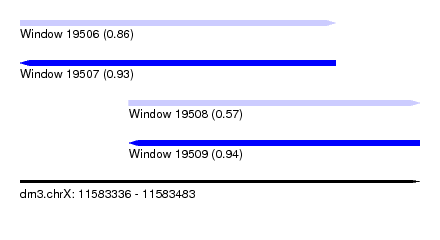
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,583,336 – 11,583,483 |
| Length | 147 |
| Max. P | 0.943632 |

| Location | 11,583,336 – 11,583,452 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Shannon entropy | 0.30583 |
| G+C content | 0.62027 |
| Mean single sequence MFE | -51.92 |
| Consensus MFE | -36.97 |
| Energy contribution | -38.28 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
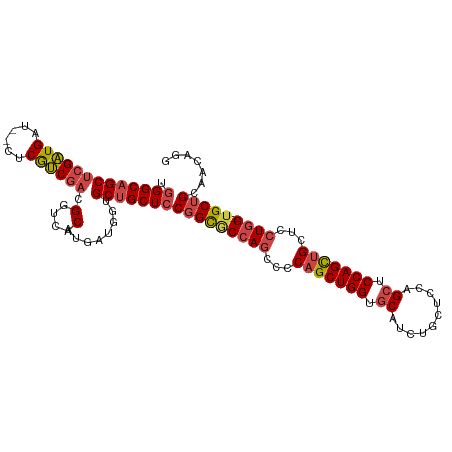
>dm3.chrX 11583336 116 + 22422827 UGAGCAAACACCAUCAGAUGCUGCUCGCCGAGCAGAUGCAUCUGUUGCAGCA-CCAGGAGCAGCUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCGCCGGAGCAGCACCAUCAU ................(.(((((((..(((.((...((((((((((.((((.-((((......)))).)))).))))))))))(((((....))))).)))))))))))).)..... ( -57.10, z-score = -2.94, R) >droSim1.chrX 8910909 116 + 17042790 UGAGCAAACACCAUCAGAUGCUGCUCGCCGAGCAGAUGCACCUGUUGCAGCA-CCAGGAGCAGCUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCGCCGGAGCAGCACCAUCAU ................(.(((((((..(((.((...((((.(((((.((((.-((((......)))).)))).))))).))))(((((....))))).)))))))))))).)..... ( -52.50, z-score = -1.70, R) >droSec1.super_36 271991 116 + 449511 UGAGCAAACACCAUCAGAUGCUGCUCGCCGAGCAGAUGCACCUGUUGCAGCA-CCAGGAGCAGCUGGACCUGGAGCAGAUGCACCAGCUGGGACUGGCGCCGGAGCAGCACCAUCAU ................(.((((((((.((.(((.(.((((.(((((.(((..-((((......))))..))).))))).)))).).))).))..((....)))))))))).)..... ( -44.20, z-score = -0.47, R) >droYak2.chrX 6668295 116 - 21770863 UGAGCAAACACCACCAGAUGCUGCUCGCCGAGCAGAUGCACCUGUUGCAGCA-CCAGGAGCAACUGGAGCUGGAGCAAAUGCACCAGCUGGGGCUGGUGCCGGAGCAGCAUCAUCAU ................(((((((((..(((.(((..((((..((((.((((.-((((......)))).)))).))))..))))(((((....))))))))))))))))))))..... ( -55.50, z-score = -3.34, R) >droEre2.scaffold_4690 15145315 116 - 18748788 UGAGCAAACACCACCAGAUGCUGCUCGCCGAGCAGAUGCACCUGUUGCAGCA-CCAGGAGCAACUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCACCGGAGCAGCAUCAUCAC ................(((((((((((((.(((...((((.(((((.((((.-((((......)))).)))).))))).))))((....)).))))))....))))))))))..... ( -55.10, z-score = -3.45, R) >anoGam1.chrX 12598409 103 - 22145176 GCAGCAGUGCGCCCCAGGUGCUGCUC---GAGCAUGCGCAGG----ACAGCUUCCAGAAGCAGCUGGAGCUUGCGCAGCAGCGCCAGCAGCAGAUUGCGCAGGAGCGGCA------- ((.((..(((((...(..(((((((.---(.((.((((((((----.(((((.(.....).)))))...))))))))...)).).)))))))..).)))))...)).)).------- ( -47.10, z-score = 0.30, R) >consensus UGAGCAAACACCACCAGAUGCUGCUCGCCGAGCAGAUGCACCUGUUGCAGCA_CCAGGAGCAGCUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCGCCGGAGCAGCACCAUCAU ..................((((((((.((.(((.(.((((.(((((.(((...((((......))))..))).))))).)))).).))).))..((....))))))))))....... (-36.97 = -38.28 + 1.31)

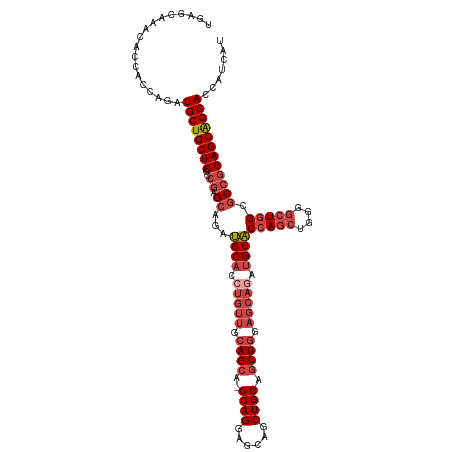
| Location | 11,583,336 – 11,583,452 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Shannon entropy | 0.30583 |
| G+C content | 0.62027 |
| Mean single sequence MFE | -55.27 |
| Consensus MFE | -41.41 |
| Energy contribution | -42.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11583336 116 - 22422827 AUGAUGGUGCUGCUCCGGCGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGCUGCUCCUGG-UGCUGCAACAGAUGCAUCUGCUCGGCGAGCAGCAUCUGAUGGUGUUUGCUCA ....((((((((...))))))))(((..(((.((((((((((...((((.((((......))))-.))))...)))))))))).))).)))((((((((((....)))).)))))). ( -61.10, z-score = -3.56, R) >droSim1.chrX 8910909 116 - 17042790 AUGAUGGUGCUGCUCCGGCGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGCUGCUCCUGG-UGCUGCAACAGGUGCAUCUGCUCGGCGAGCAGCAUCUGAUGGUGUUUGCUCA ....((((((((...))))))))(((..(((.((((((((((...((((.((((......))))-.))))...)))))))))).))).)))((((((((((....)))).)))))). ( -60.50, z-score = -3.03, R) >droSec1.super_36 271991 116 - 449511 AUGAUGGUGCUGCUCCGGCGCCAGUCCCAGCUGGUGCAUCUGCUCCAGGUCCAGCUGCUCCUGG-UGCUGCAACAGGUGCAUCUGCUCGGCGAGCAGCAUCUGAUGGUGUUUGCUCA ....((((((((...))))))))(.((.(((.((((((((((...(((..((((......))))-..)))...)))))))))).))).)))((((((((((....)))).)))))). ( -54.10, z-score = -1.47, R) >droYak2.chrX 6668295 116 + 21770863 AUGAUGAUGCUGCUCCGGCACCAGCCCCAGCUGGUGCAUUUGCUCCAGCUCCAGUUGCUCCUGG-UGCUGCAACAGGUGCAUCUGCUCGGCGAGCAGCAUCUGGUGGUGUUUGCUCA .....((((((((((.(((....)))(((((.((((((((((...((((.((((......))))-.))))...)))))))))).))).)).))))))))))................ ( -54.00, z-score = -1.99, R) >droEre2.scaffold_4690 15145315 116 + 18748788 GUGAUGAUGCUGCUCCGGUGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGUUGCUCCUGG-UGCUGCAACAGGUGCAUCUGCUCGGCGAGCAGCAUCUGGUGGUGUUUGCUCA .....((((((((((.(....).(((..(((.((((((((((...((((.((((......))))-.))))...)))))))))).))).)))))))))))))................ ( -55.10, z-score = -2.00, R) >anoGam1.chrX 12598409 103 + 22145176 -------UGCCGCUCCUGCGCAAUCUGCUGCUGGCGCUGCUGCGCAAGCUCCAGCUGCUUCUGGAAGCUGU----CCUGCGCAUGCUC---GAGCAGCACCUGGGGCGCACUGCUGC -------.((.((...(((((....(((((((.(.((...((((((((((((((......)))).))))..----..)))))).)).)---.)))))))......)))))..)).)) ( -46.80, z-score = 0.08, R) >consensus AUGAUGGUGCUGCUCCGGCGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGCUGCUCCUGG_UGCUGCAACAGGUGCAUCUGCUCGGCGAGCAGCAUCUGAUGGUGUUUGCUCA .....((((((((((.(((....)))(((((.((((((((((...(((..((((......))))...)))...)))))))))).))).)).))))))))))................ (-41.41 = -42.42 + 1.00)

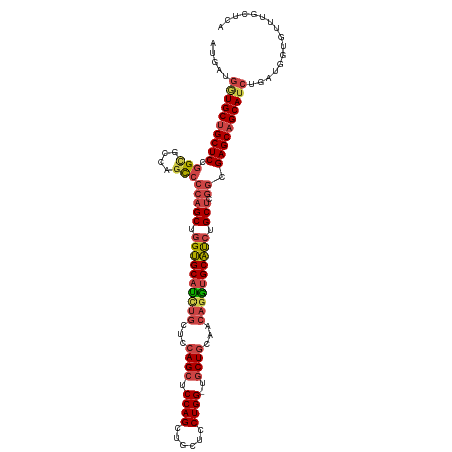
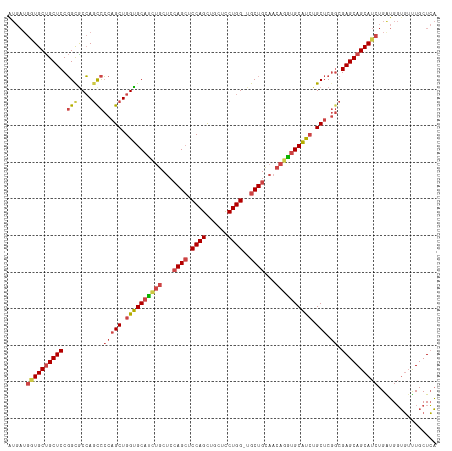
| Location | 11,583,376 – 11,583,483 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.83 |
| Shannon entropy | 0.31029 |
| G+C content | 0.63870 |
| Mean single sequence MFE | -48.33 |
| Consensus MFE | -32.45 |
| Energy contribution | -33.65 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.574140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11583376 107 + 22422827 UCUGUUGCAGCA-CCAGGAGCAGCUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCGCCGGAGCAGCACCAUCAUGUGACCGUCGACGAG--AUCAUCGAGCUGCCCA ((((((.((((.-((((......)))).)))).)))))).((.(((((....))))).)).((.(((((....((....))...((((.(..--..).))))))))))). ( -48.30, z-score = -1.43, R) >droSim1.chrX 8910949 107 + 17042790 CCUGUUGCAGCA-CCAGGAGCAGCUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCGCCGGAGCAGCACCAUCAUGUGACCGUCGACGAG--AUCAUCGAGCUGCCCA .(((((.((((.-((((......)))).)))).)))))..((.(((((....))))).)).((.(((((....((....))...((((.(..--..).))))))))))). ( -47.20, z-score = -1.11, R) >droSec1.super_36 272031 107 + 449511 CCUGUUGCAGCA-CCAGGAGCAGCUGGACCUGGAGCAGAUGCACCAGCUGGGACUGGCGCCGGAGCAGCACCAUCAUGUGACCGUCGACGAG--AUCAUCGAGCUGCCCA .(((((.(((..-((((......))))..))).)))))..((.((((......)))).)).((.(((((....((....))...((((.(..--..).))))))))))). ( -38.50, z-score = 0.39, R) >droYak2.chrX 6668335 107 - 21770863 CCUGUUGCAGCA-CCAGGAGCAACUGGAGCUGGAGCAAAUGCACCAGCUGGGGCUGGUGCCGGAGCAGCAUCAUCAUGUGACCGUCGACGAG--AUCAUCGAGCUGCCCA ..((((.((((.-((((......)))).)))).))))...((((((((....)))))))).((.(((((.((((...))))...((((.(..--..).))))))))))). ( -50.10, z-score = -2.77, R) >droEre2.scaffold_4690 15145355 107 - 18748788 CCUGUUGCAGCA-CCAGGAGCAACUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCACCGGAGCAGCAUCAUCACGUCACCGUCGACGAG--AUCAUCGAGCUGCCCA .(((((.((((.-((((......)))).)))).))))).....(((((....)))))....((.(((((.(((((.((((......)))).)--))....))))))))). ( -48.00, z-score = -2.32, R) >anoGam1.chrX 12598446 104 - 22145176 ----GGACAGCUUCCAGAAGCAGCUGGAGCUUGCGCAGCAGCGCCAGCAGCAGAUUGCGCAGGAGCGGCAGCGAC-UGCAGGCGCUGCUGCCCCAUCAGCG-GCUGCCAC ----((.((((((((((......)))))(((((.(((((((((((.((((....))))((((..((....))..)-))).)))))))))))..))..))))-)))))).. ( -57.90, z-score = -2.61, R) >consensus CCUGUUGCAGCA_CCAGGAGCAGCUGGAGCUGGAGCAGAUGCACCAGCUGGGGCUGGCGCCGGAGCAGCACCAUCAUGUGACCGUCGACGAG__AUCAUCGAGCUGCCCA .(((((.(((...((((......))))..))).)))))..((.(((((....))))).)).((.(((((.......((....))((((.((....)).))))))))))). (-32.45 = -33.65 + 1.20)

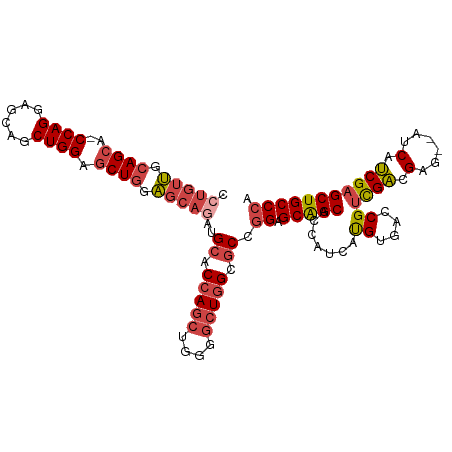
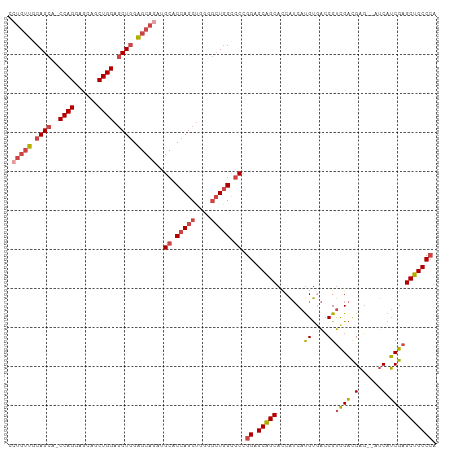
| Location | 11,583,376 – 11,583,483 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Shannon entropy | 0.31029 |
| G+C content | 0.63870 |
| Mean single sequence MFE | -51.88 |
| Consensus MFE | -37.03 |
| Energy contribution | -37.82 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11583376 107 - 22422827 UGGGCAGCUCGAUGAU--CUCGUCGACGGUCACAUGAUGGUGCUGCUCCGGCGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGCUGCUCCUGG-UGCUGCAACAGA .(((((((.(((((..--..)))))((.(((....))).)))))))))(((((((((...(((((((.((..........)).)))))))...))))-)))))....... ( -51.90, z-score = -2.30, R) >droSim1.chrX 8910949 107 - 17042790 UGGGCAGCUCGAUGAU--CUCGUCGACGGUCACAUGAUGGUGCUGCUCCGGCGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGCUGCUCCUGG-UGCUGCAACAGG .(((((((.(((((..--..)))))((.(((....))).)))))))))(((((((((...(((((((.((..........)).)))))))...))))-)))))....... ( -51.90, z-score = -2.25, R) >droSec1.super_36 272031 107 - 449511 UGGGCAGCUCGAUGAU--CUCGUCGACGGUCACAUGAUGGUGCUGCUCCGGCGCCAGUCCCAGCUGGUGCAUCUGCUCCAGGUCCAGCUGCUCCUGG-UGCUGCAACAGG .(((((((.(((((..--..)))))((.(((....))).)))))))))(((((((((...(((((((...(((((...))))))))))))...))))-)))))....... ( -50.90, z-score = -2.06, R) >droYak2.chrX 6668335 107 + 21770863 UGGGCAGCUCGAUGAU--CUCGUCGACGGUCACAUGAUGAUGCUGCUCCGGCACCAGCCCCAGCUGGUGCAUUUGCUCCAGCUCCAGUUGCUCCUGG-UGCUGCAACAGG .(((((((((((((..--..))))))..((((.....)))))))))))(((((((((...(((((((.((..........)).)))))))...))))-)))))....... ( -48.40, z-score = -2.46, R) >droEre2.scaffold_4690 15145355 107 + 18748788 UGGGCAGCUCGAUGAU--CUCGUCGACGGUGACGUGAUGAUGCUGCUCCGGUGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGUUGCUCCUGG-UGCUGCAACAGG .(((((((.(((((..--..)))))(((....)))......))))).)).(..(((((....)))))..)..(((...((((.((((......))))-.))))...))). ( -49.90, z-score = -2.25, R) >anoGam1.chrX 12598446 104 + 22145176 GUGGCAGC-CGCUGAUGGGGCAGCAGCGCCUGCA-GUCGCUGCCGCUCCUGCGCAAUCUGCUGCUGGCGCUGCUGCGCAAGCUCCAGCUGCUUCUGGAAGCUGUCC---- ..((((((-.(((....(.(((((((((((.(((-((.(.(((.((....)))))..).))))).))))))))))).).)))(((((......))))).)))))).---- ( -58.30, z-score = -1.97, R) >consensus UGGGCAGCUCGAUGAU__CUCGUCGACGGUCACAUGAUGGUGCUGCUCCGGCGCCAGCCCCAGCUGGUGCAUCUGCUCCAGCUCCAGCUGCUCCUGG_UGCUGCAACAGG .(((((((((((((......)))))).(....)........)))))))(((((((((...(((((((.((..........)).)))))))...)))).)))))....... (-37.03 = -37.82 + 0.78)
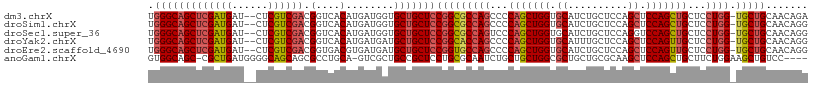
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:10 2011