| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,077,622 – 11,077,720 |
| Length | 98 |
| Max. P | 0.870083 |

| Location | 11,077,622 – 11,077,720 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 69.73 |
| Shannon entropy | 0.66421 |
| G+C content | 0.46781 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -11.46 |
| Energy contribution | -12.49 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
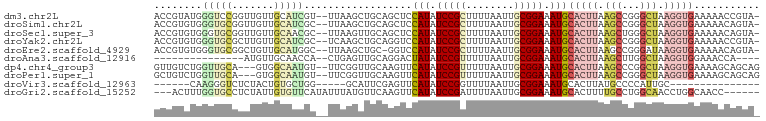
>dm3.chr2L 11077622 98 - 23011544 ACCGUAUGGGUCCGGUUGUUGCAUCGU--UUAAGCUGCAGCUCCAUAUCCGCUUUUAAUUGCGGAAAUGCACUUAAGCCGGGCUAAGGUGAAAAACCGUA- ........(((((((((((((((.(..--....).))))))..(((.(((((........))))).)))......)))))))))..(((.....)))...- ( -35.60, z-score = -2.70, R) >droSim1.chr2L 10876648 98 - 22036055 ACCGUGUGGGUGCGGUUGUUGCAUCGC--UUAAGCUGCAGCUCCAUAUCCGCUUUUAAUUGCGGAAAUGCACUUAAGCCGGGCUAAGGUGAAAAACAGUA- .(((..(((((((((..((((((..((--....)))))))).))...(((((........)))))...)))))))...)))...................- ( -33.50, z-score = -1.31, R) >droSec1.super_3 6479076 98 - 7220098 ACCGUGUGGGUGCGGUUGUUGCAACGC--UUAAGUUGCAGCUCCAUAUCCGCUUUUAAUUGCGGAAAUGCACUUAAGCUGGGCUAAGGUGAAAAACAGUA- .((((.(((((((((..((((((((..--....)))))))).))...(((((........)))))...))))))).)).))...................- ( -36.10, z-score = -2.53, R) >droYak2.chr2L 7473612 98 - 22324452 ACCGUGUGGGUGCGCUUGUUGCAUCGC--UCAAGCUGCAGGUCCAUAUCCGCUUUUAAUUGCGGAAAUGCACUUAAGCCGGGCUAAGGUGAAAAACCGUA- .(((..(((((((((((((.((.....--....)).)))))).....(((((........)))))...)))))))...))).....(((.....)))...- ( -32.80, z-score = -0.92, R) >droEre2.scaffold_4929 12274229 97 + 26641161 ACCGUGUGGGUGCGGCUGUUGCAUGGC--UUAAGCUGC-GGUCCAUAUCCGCUUUUAAUUGCGGAAAUGCACUUAAGCCGGGAUAAGGUGAAAAACAGUA- .((((......))))(((((.((((((--(((((.(((-(.......(((((........)))))..))))))))))))........)))...)))))..- ( -31.60, z-score = -0.70, R) >droAna3.scaffold_12916 12202577 80 - 16180835 ---------------AUGUUGCAACCA--CUGAGUUGCAGGACUAUAUCCGUUUUUAAUUGCGGAAAUGCACUUAAGCUUGGCUAAGGUGGAAACCA---- ---------------....((((((..--....))))))((......(((((........)))))..(.(((((.(((...))).))))).)..)).---- ( -22.10, z-score = -1.42, R) >dp4.chr4_group3 6422833 96 - 11692001 GUUGUCUGGUUGCA---GUGGCAAUGU--UUCGGUUGCAAGUUCAUAUCCGUUUUUAAUUGCGGAAAUGCACUUAAGCCCGGCUAAGGUGAAAAGCAGCAG ((((.((..(..(.---.((((.....--...((((..((((.(((.(((((........))))).))).)))).))))..))))..)..)..)))))).. ( -30.00, z-score = -1.71, R) >droPer1.super_1 7913961 96 - 10282868 GCUGUCUGGUUGCA---GUGGCAAUGU--UUCGGUUGCAAGUUCAUAUCCGUUUUUAAUUGCGGAAAUGCACUUAAGCCGGGCUAAGGUGAAAAGCAGCAG ((((.((..(..(.---.((((.....--.((((((..((((.(((.(((((........))))).))).)))).))))))))))..)..)..)))))).. ( -35.80, z-score = -3.28, R) >droVir3.scaffold_12963 12240226 75 + 20206255 ------CAAGGGUCUCUACUGUGCUGG-----GCAUUCGAGUUCAUAUCCGGUUUUAAUUGCGGAAAUGCACUUAUGCCCCAUUGC--------------- ------...................((-----((((..((((.(((.((((..........)))).))).))))))))))......--------------- ( -19.20, z-score = -0.48, R) >droGri2.scaffold_15252 6156883 92 + 17193109 ---ACUUUGGUGCCUCUAUUGUGUUCAUAUUUAUGUUCAAGUUCAUAUCCGAUUUUAAUUGCGGAAAUGCACUUUUGCCUGGCAACCUGGCAACC------ ---.....(((((........((..(((....)))..))........((((..........))))...))))).(((((.(....)..)))))..------ ( -18.40, z-score = -0.39, R) >consensus ACCGUGUGGGUGCGGUUGUUGCAUCGC__UUAAGCUGCAGGUCCAUAUCCGCUUUUAAUUGCGGAAAUGCACUUAAGCCGGGCUAAGGUGAAAAACAGUA_ ........(((((.......)))))..................(((.(((((........))))).)))(((((.(((...))).)))))........... (-11.46 = -12.49 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:31 2011