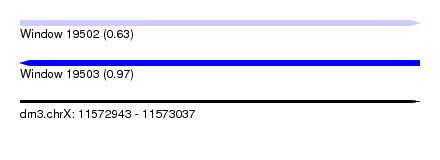
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,572,943 – 11,573,037 |
| Length | 94 |
| Max. P | 0.970329 |

| Location | 11,572,943 – 11,573,037 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.50938 |
| G+C content | 0.45194 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -12.82 |
| Energy contribution | -14.69 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626856 |
| Prediction | RNA |
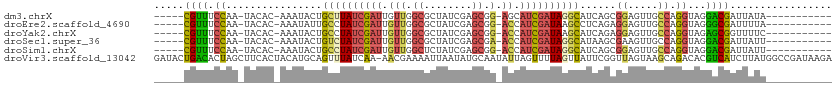
Download alignment: ClustalW | MAF
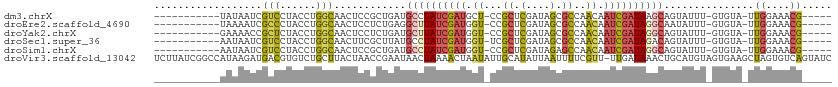
>dm3.chrX 11572943 94 + 22422827 -----------UAUAAUCGUCCUACCUGGCAACUCCGCUGAUGCCUAUCGAUGCU-CCGCUCGAUAGCGCCAACAAUCGAUAAGCAGUAUUU-GUGUA-UUGGAAACG----- -----------....((((.......((((......((....))(((((((.((.-..))))))))).)))).....))))...((((((..-..)))-)))......----- ( -21.70, z-score = -0.80, R) >droEre2.scaffold_4690 15135295 94 - 18748788 -----------UAAAAUCGCCCUACCUGGCAACUCCUCUGAGGCUUAUCGAUGGU-CCGCUCGAUAGCGCCAACAAUCGAUAGGCAAUAUUU-GUGUA-UUGGAAACG----- -----------.......(((......)))..(((....)))((((((((((.((-.((((....))))...)).))))))))))(((((..-..)))-))(....).----- ( -26.00, z-score = -1.80, R) >droYak2.chrX 6657954 94 - 21770863 -----------GAAAACCGCUCUACCUGGCAACUCCUCUGAUGCUUAUCGAUGGU-CCGCUCGAUAGCGCCAACAAUCGAUAGGCAGUAUUU-GUGUA-UUGGAAACG----- -----------......(((..(((..(....)........(((((((((((.((-.((((....))))...)).))))))))))))))...-)))..-..(....).----- ( -23.90, z-score = -1.10, R) >droSec1.super_36 261691 94 + 449511 -----------AAUAAUCGUCCUACCUGGCAACUUCGCUUAUGCCUAUCGAUGGU-UCGCUCGAUAGCGCCAACAAUCGAUAGACAGUAUUU-GUGUA-UUGGAAACG----- -----------........(((.....(....)..(((..((((((((((((.((-(((((....))))..))).))))))))...))))..-)))..-..)))....----- ( -22.80, z-score = -1.10, R) >droSim1.chrX 8899845 94 + 17042790 -----------AAUAAUCGUCCUACCUGGCAACUCCGCUGAUGCCUAUCGAUGGU-CCGCUCGAUAGAGCCAACAAUCGAUAGGCAGUAUUU-GUGUA-UUGGAAACG----- -----------........(((.....(((......)))..(((((((((((.((-..((((....))))..)).)))))))))))......-.....-..)))....----- ( -30.80, z-score = -3.09, R) >droVir3.scaffold_13042 2779966 112 + 5191987 UCUUAUCGGCCAUAAGAUGACGUGUCUGCUUACUAACCGAAUAACUAAAACUAAUAUUGCAUAUUAAUUUUCGUU-UUGAUAAACUGCAUGUAGUGAAGCUAGUGUCAGUAUC ((((((.....))))))(((((((......)))..........((((..((((....((((((((((........-))))))...))))..)))).....))))))))..... ( -16.80, z-score = 0.80, R) >consensus ___________AAAAAUCGUCCUACCUGGCAACUCCGCUGAUGCCUAUCGAUGGU_CCGCUCGAUAGCGCCAACAAUCGAUAGGCAGUAUUU_GUGUA_UUGGAAACG_____ ........................((..(((..........(((((((((((.((..(((......)))...)).)))))))))))........)))....)).......... (-12.82 = -14.69 + 1.87)
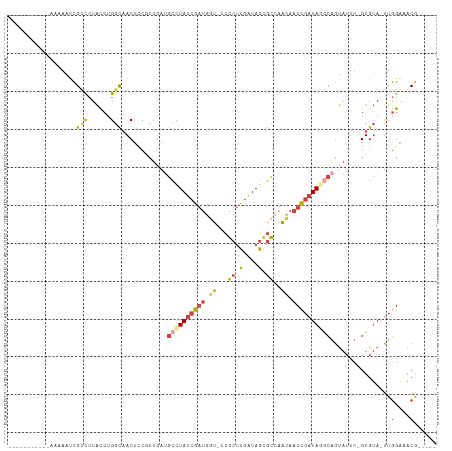
| Location | 11,572,943 – 11,573,037 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.50938 |
| G+C content | 0.45194 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -16.20 |
| Energy contribution | -13.72 |
| Covariance contribution | -2.48 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
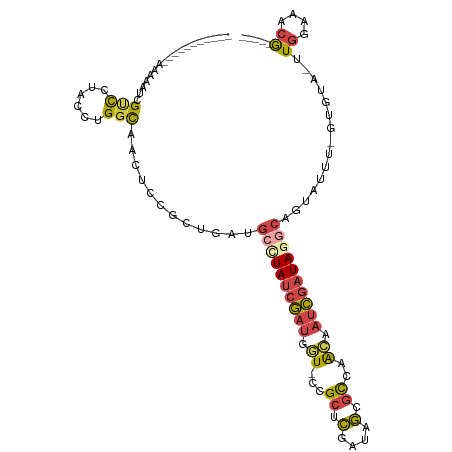
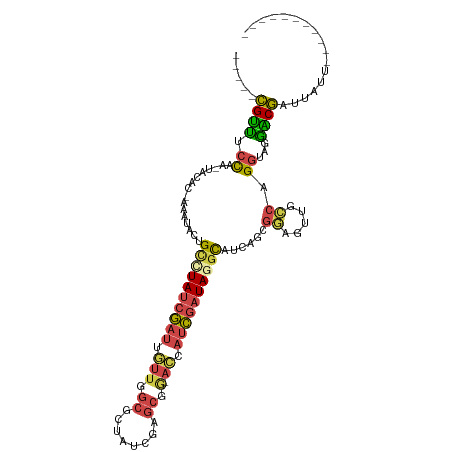
>dm3.chrX 11572943 94 - 22422827 -----CGUUUCCAA-UACAC-AAAUACUGCUUAUCGAUUGUUGGCGCUAUCGAGCGG-AGCAUCGAUAGGCAUCAGCGGAGUUGCCAGGUAGGACGAUUAUA----------- -----((((.((..-.....-......(((((((((((.(((..((((....)))).-)))))))))))))).....((.....)).))...))))......----------- ( -26.30, z-score = -0.98, R) >droEre2.scaffold_4690 15135295 94 + 18748788 -----CGUUUCCAA-UACAC-AAAUAUUGCCUAUCGAUUGUUGGCGCUAUCGAGCGG-ACCAUCGAUAAGCCUCAGAGGAGUUGCCAGGUAGGGCGAUUUUA----------- -----.........-.....-.......((.(((((((.(((.((........)).)-)).))))))).))......(((((((((......))))))))).----------- ( -26.90, z-score = -1.19, R) >droYak2.chrX 6657954 94 + 21770863 -----CGUUUCCAA-UACAC-AAAUACUGCCUAUCGAUUGUUGGCGCUAUCGAGCGG-ACCAUCGAUAAGCAUCAGAGGAGUUGCCAGGUAGAGCGGUUUUC----------- -----(((((((..-.....-......(((.(((((((.(((.((........)).)-)).))))))).))).....((.....)).))..)))))......----------- ( -25.00, z-score = -0.82, R) >droSec1.super_36 261691 94 - 449511 -----CGUUUCCAA-UACAC-AAAUACUGUCUAUCGAUUGUUGGCGCUAUCGAGCGA-ACCAUCGAUAGGCAUAAGCGAAGUUGCCAGGUAGGACGAUUAUU----------- -----((((.((..-.....-......(((((((((((.(((.((........)).)-)).)))))))))))...(((....)))..))...))))......----------- ( -27.50, z-score = -2.27, R) >droSim1.chrX 8899845 94 - 17042790 -----CGUUUCCAA-UACAC-AAAUACUGCCUAUCGAUUGUUGGCUCUAUCGAGCGG-ACCAUCGAUAGGCAUCAGCGGAGUUGCCAGGUAGGACGAUUAUU----------- -----((((.((..-.....-......(((((((((((.(((.((((....)))).)-)).))))))))))).....((.....)).))...))))......----------- ( -34.80, z-score = -4.02, R) >droVir3.scaffold_13042 2779966 112 - 5191987 GAUACUGACACUAGCUUCACUACAUGCAGUUUAUCAA-AACGAAAAUUAAUAUGCAAUAUUAGUUUUAGUUAUUCGGUUAGUAAGCAGACACGUCAUCUUAUGGCCGAUAAGA (((((((....(((.....)))....))))..)))..-(((.(((((((((((...))))))))))).)))..(((((((.((((..((....))..)))))))))))..... ( -22.00, z-score = -1.25, R) >consensus _____CGUUUCCAA_UACAC_AAAUACUGCCUAUCGAUUGUUGGCGCUAUCGAGCGG_ACCAUCGAUAGGCAUCAGCGGAGUUGCCAGGUAGGACGAUUAUU___________ .....((((.................((((((..(((((.((((.((((((((..(....).))))).))).))))...)))))..))))))))))................. (-16.20 = -13.72 + -2.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:05 2011