| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,567,231 – 11,567,327 |
| Length | 96 |
| Max. P | 0.799529 |

| Location | 11,567,231 – 11,567,327 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Shannon entropy | 0.54611 |
| G+C content | 0.45666 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -15.76 |
| Energy contribution | -15.50 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
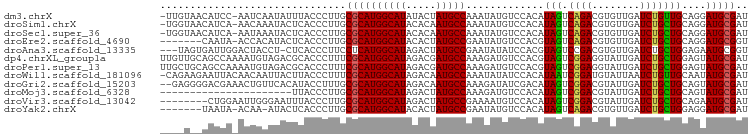
>dm3.chrX 11567231 96 - 22422827 -UUGUAACAUCC-AAUCAAUAUUUACCCUUGCGCAUGGCAUAUACUAUGCCAAAUAUGUCCACAUAGUCAGACGUGUUGAUCUGUUGCAGGAUGCGAU -((((((((...-.((((((((......(((.((((((......))))))))).((((....)))).......)))))))).))))))))........ ( -22.30, z-score = -0.61, R) >droSim1.chrX 8896146 96 - 17042790 -UGGUAACAUCA-AACAAAUACUCACCCUUGCGCAUGGCAUACACAAUGCCAAAUAUGUCCACAUAGUCAGACGUGUUGAUCUGCUGCAGGAUGCGAU -...........-........(.((.(((.(((((((((((.....))))))((((((((..........))))))))....))).))))).)).).. ( -22.60, z-score = -0.16, R) >droSec1.super_36 256021 96 - 449511 -UGGUAACAUCA-AAUAAAUACUCACCCUUGCGCAUGGCAUACACAAUGCCAAAUAUGUCCACAUAGUCAGACGUGUUGAUCUGCUGCAGGAUGCGAU -...........-........(.((.(((.(((((((((((.....))))))((((((((..........))))))))....))).))))).)).).. ( -22.60, z-score = -0.23, R) >droEre2.scaffold_4690 15129571 90 + 18748788 -------CAAUA-ACCACAUACUCACCCUUGCGCAUGGCAUACACUAUGCCGAAUAUGUCCACGUAGUCAGACGUGUUGAUCUGCUGCAGGAUGCGGU -------.....-(((.(((.......(((((((((((((((...)))))))((((((((..........))))))))....))).)))))))).))) ( -24.51, z-score = -0.39, R) >droAna3.scaffold_13335 2820094 94 + 3335858 ---UAGUGAUUGGACUACCU-CUCACCCUUCCUCAUGGCAUAGACUAUGCCGAAUAUAUCCACGUAGUCCGACGUGUUGAUCUGCUGGAGAAUGCGGU ---..((((..((....)).-.))))....((.(((((((((...)))))).......((((.((((..(((....)))..))))))))..))).)). ( -26.10, z-score = -1.05, R) >dp4.chrXL_group1a 7301946 98 - 9151740 UUGUUGCAGCCAAAAUGUAGACGCACCCUUUCGCAUGGCAUAGACGAUGCCAAAGAUGUCCACGUAGUCGGAGGUAUUGAUCUGCUGGAGUAUGCGAU ...(((((.......))))).((((..(((..(((((((((.....))))))..(((..(..((....))..)..)))....)))..)))..)))).. ( -26.70, z-score = -0.23, R) >droPer1.super_13 1920059 98 - 2293547 UUGCUGCAGCCAAAAUGUAGACGCACCCUUUCGCAUGGCAUAGACGAUGCCAAAGAUGUCCACGUAGUCGGAGGUAUUGAUCUGCUGGAGUAUGCGAU ...(((((.......))))).((((..(((..(((((((((.....))))))..(((..(..((....))..)..)))....)))..)))..)))).. ( -29.00, z-score = -0.67, R) >droWil1.scaffold_181096 5838035 97 + 12416693 -CAGAAGAAUUACAACAAUUACUUACCCUUUCGCAUGGCAUAGACAAUGCCAAAUAUAUCCACAUAAUCGGAUGUAUUAAUCUGUUGCAAUAUGCGAU -.............................(((((((((((.....))))).(((((((((........))))))))).............)))))). ( -22.90, z-score = -3.18, R) >droGri2.scaffold_15203 5555306 96 - 11997470 --GAGGGGACGAAACUGUUCACAUACCUUUGCGCAUGGCAUAGACAAUGCCAAAGAUAUCGACAUAGUCGGACGUAUUGAUCUGCUGCAGUAUGCGAU --..(....)........((.(((((...((((((((((((.....))))))......(((((...)))))...........))).)))))))).)). ( -23.90, z-score = -0.30, R) >droMoj3.scaffold_6328 3611517 76 + 4453435 ----------------------UUACCCUUGCGCAUGGCAUAGACUAUGCCAAAGAUGUCCACAUAGUCGGACGUAUUGAUCUGCUGCAGUAUGCGAU ----------------------......((((((((((((((...)))))))...((((((........)))))).......))).))))........ ( -20.90, z-score = -0.94, R) >droVir3.scaffold_13042 2773093 90 - 5191987 --------CUGGAAUUGGGAAUUUACCCUUGCGCAUGGCAUAGACUAUGCCGAAAAUGUCCACAUAGUCGGACGUAUUGAUCUGCUGCAGAAUGCGAU --------........(((......)))...(((((.(((((((.((((((((..(((....)))..)))).))))....)))).)))...))))).. ( -26.30, z-score = -1.64, R) >droYak2.chrX 6652147 89 + 21770863 -------UAAUA-ACAA-AUACUCACCCUUGCGCAUGGCAUACACUAUGCCGAAUAUGUCCACAUAGUCAGACGUGUUGAUCUGCUGGAGGAUGCGAU -------.....-....-...(.((.((((..((((((((((...)))))))((((((((..........))))))))....)))..)))).)).).. ( -21.20, z-score = -0.61, R) >consensus ____U__CAUCA_AAUAAAUACUCACCCUUGCGCAUGGCAUAGACUAUGCCAAAUAUGUCCACAUAGUCGGACGUAUUGAUCUGCUGCAGGAUGCGAU ...............................((((((((((.....))))).............(((.((((........)))))))....))))).. (-15.76 = -15.50 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:02 2011