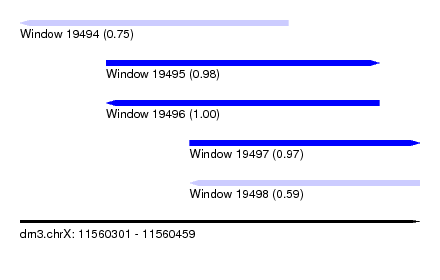
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,560,301 – 11,560,459 |
| Length | 158 |
| Max. P | 0.998244 |

| Location | 11,560,301 – 11,560,407 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.43 |
| Shannon entropy | 0.49217 |
| G+C content | 0.48845 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -9.34 |
| Energy contribution | -10.03 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
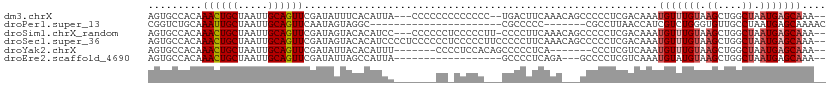
>dm3.chrX 11560301 106 - 22422827 AGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUUCACAUUA---CCCCCCCCCCCCC--UGACUUCAAACAGCCCCCUCGACAAAUGUUUGUAAGCUGGCUAAUGAGCAAA-- ..((((((.((((((.....)))))).).............---...........((--.(.(((((((((..............)))))).)))).))....)).)))..-- ( -18.14, z-score = -1.53, R) >droPer1.super_13 382510 84 + 2293547 CGGUCUGCAAAUUGCUAAUUGCAGUUCAAUAGUAGGC----------------------CGCCCCC-------CGCCUUAACCAUCGUCUGGGUGUUGCCUAAUGAGCAAAAC ((((((((.((((((.....)))))).....))))))----------------------)).....-------(((((..((....))..)))))(((((....).))))... ( -24.80, z-score = -1.84, R) >droSim1.chrX_random 3233091 107 - 5698898 AGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAUCC---CCCCCCUCCCCCUU-CCCCUUCAAACAGCCCCCUCGACAAAUGUUUGUAAGCUGGCUAAUGAGCAAA-- .((((..(.((((((.....)))))).)...))))......---..............-....((((...((((..((..((((....)))).))..))))..))))....-- ( -19.30, z-score = -2.14, R) >droSec1.super_36 248404 111 - 449511 AGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAUCCCCUCCCCCCUCCCCCUUCCCCCUUCAAACAGCCCCCUCGACAAAUGUUUGUAAGCUGGCUAAUGAGCAAA-- .((((..(.((((((.....)))))).)...))))..............(((......((..(((((((((..............)))))).)))..)).....)))....-- ( -19.54, z-score = -2.27, R) >droYak2.chrX 6645369 97 + 21770863 AGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUACACAUUU-------CCCCUCCACAGCCCCCUCA-------CCCUCGUCAAAUGUUUGUAAGCUGGCUAAUGAGCAAA-- ..((((((.((((((.....)))))).).............-------.........((((..((.(-------(...(((...)))...)).))..))))..)).)))..-- ( -16.90, z-score = -0.92, R) >droEre2.scaffold_4690 15122917 90 + 18748788 AGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUAGCCAUUA------------------GCCCCUCAGA---GCCCCUCGUCAAAUGUAUGUAAGCUGGCUAAUGAGCAAA-- .......(.((((((.....)))))).)......(((((((------------------(((......(---((....((((....).)))...))))))))))).))...-- ( -22.80, z-score = -1.87, R) >consensus AGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAUUA_______CC_CCCCC___CCCCCUCAAA___CCCCCUCGACAAAUGUUUGUAAGCUGGCUAAUGAGCAAA__ .........((((((.....))))))...........................................................(((((((.((....)).))))))).... ( -9.34 = -10.03 + 0.70)
| Location | 11,560,335 – 11,560,443 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.59 |
| Shannon entropy | 0.51974 |
| G+C content | 0.49764 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -18.12 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11560335 108 + 22422827 GGGGGCUGUUUGAAGUCAG-----GGGGGGGGGGGGUAAUGUGAAAUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAU--GUUUCGUUUGCUCG- ...((((......))))..-----(((.(((.((..((..(((...((.((((((((.....)))))))))).))).((((((.((....)))))))))--)..)).))).))).- ( -33.50, z-score = -2.16, R) >droPer1.super_13 382546 87 - 2293547 -----------------------------GGCGGGGGGCGGCCUACUAUUGAACUGCAAUUAGCAAUUUGCAGACCGUUCGUCGUGUAUUUGGUUUCAUCGAUAGAGUUUUCUCUC -----------------------------...((((((..((...(((((((.((((((........))))))((((.............))))....))))))).))..)))))) ( -22.62, z-score = -0.52, R) >droSim1.chrX_random 3233125 110 + 5698898 GGGGGCUGUUUGAAGGGGAA----GGGGGAGGGGGGGGAUGUGUACUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAU--GUUUCGUUUGCUCUC ((((((....(((((.(...----................((((.....((((((((.....))))))))..)))).((((((.((....)))))))))--.)))))...)))))) ( -33.80, z-score = -1.69, R) >droSec1.super_36 248438 114 + 449511 GGGGGCUGUUUGAAGGGGGAAGGGGGAGGGGGGAGGGGAUGUGUACUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAU--GUUUCGUUUGCUCUC .....((.(((.......))).)).(((((.(((.(..((((((.....((((((((.....))))))))..)))).((((((.((....)))))))).--))..).))).))))) ( -36.70, z-score = -2.23, R) >droYak2.chrX 6645407 96 - 21770863 -------------GAGGGG-----GCUGUGGAGGGGAAAUGUGUAAUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACACGUAUUUGGUUGCAU--GUUUCGUUUGCUUUC -------------..(..(-----((.....(((.(((((((((.....((((((((.....))))))))..)))).((((((.((....)))))))).--))))).))))))..) ( -30.30, z-score = -2.46, R) >droEre2.scaffold_4690 15122951 93 - 18748788 ----------------GGG-----GCUCUGAGGGGCUAAUGGCUAAUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUUGGUUGCAU--GUUUCGCUUGCUUUC ----------------..(-----((((....)))))...(((.......(((((((.....)))))))((((.((.((((((.((....)))))))).--)).))))..)))... ( -29.30, z-score = -1.95, R) >consensus _____________AGGGGG_____GGGGGGGGGGGGGAAUGUGUAAUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAU__GUUUCGUUUGCUCUC ..............................(((((.(((((........((((((((.....)))))))).......((((((.((....)))))))).......))))).))))) (-18.12 = -19.02 + 0.89)
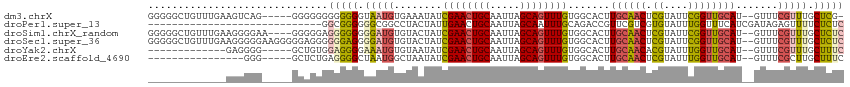
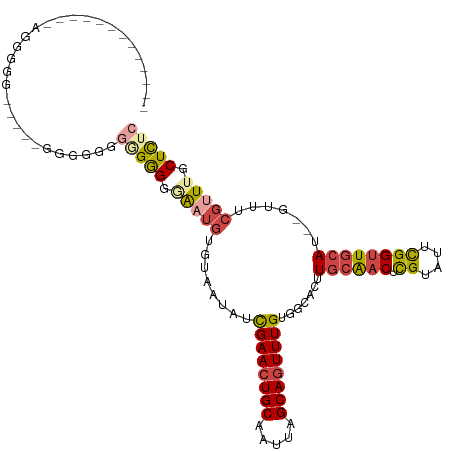
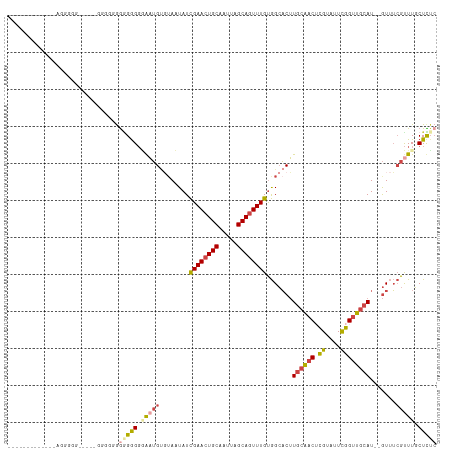
| Location | 11,560,335 – 11,560,443 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.59 |
| Shannon entropy | 0.51974 |
| G+C content | 0.49764 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -10.44 |
| Energy contribution | -11.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11560335 108 - 22422827 -CGAGCAAACGAAAC--AUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUUCACAUUACCCCCCCCCCCC-----CUGACUUCAAACAGCCCCC -...((....(..((--.((((((((....)).)))))).))..).(.((((((.....)))))).).........................-----.............)).... ( -19.70, z-score = -3.33, R) >droPer1.super_13 382546 87 + 2293547 GAGAGAAAACUCUAUCGAUGAAACCAAAUACACGACGAACGGUCUGCAAAUUGCUAAUUGCAGUUCAAUAGUAGGCCGCCCCCCGCC----------------------------- .((((....)))).(((.((..........)))))....((((((((.((((((.....)))))).....)))))))).........----------------------------- ( -19.20, z-score = -1.84, R) >droSim1.chrX_random 3233125 110 - 5698898 GAGAGCAAACGAAAC--AUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAUCCCCCCCCUCCCCC----UUCCCCUUCAAACAGCCCCC (((............--.((((((((....)).)))))).((((..(.((((((.....)))))).)...))))...........)))....----.................... ( -22.10, z-score = -4.16, R) >droSec1.super_36 248438 114 - 449511 GAGAGCAAACGAAAC--AUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAUCCCCUCCCCCCUCCCCCUUCCCCCUUCAAACAGCCCCC (((............--.((((((((....)).)))))).((((..(.((((((.....)))))).)...)))).......)))................................ ( -22.10, z-score = -4.09, R) >droYak2.chrX 6645407 96 + 21770863 GAAAGCAAACGAAAC--AUGCAACCAAAUACGUGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUACACAUUUCCCCUCCACAGC-----CCCCUC------------- ....((....(..((--.(((((((......).)))))).))..).(.((((((.....)))))).).......................))-----......------------- ( -17.90, z-score = -2.32, R) >droEre2.scaffold_4690 15122951 93 + 18748788 GAAAGCAAGCGAAAC--AUGCAACCAAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUAGCCAUUAGCCCCUCAGAGC-----CCC---------------- ((..((....(..((--.(((((((......).)))))).))..).(.((((((.....)))))).).............))...)).....-----...---------------- ( -18.00, z-score = -1.39, R) >consensus GAGAGCAAACGAAAC__AUGCAACCAAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAUCCCCCCCCCCCACC_____CCCCCU_____________ ..................(((((((......).)))))).........((((((.....))))))................................................... (-10.44 = -11.13 + 0.69)
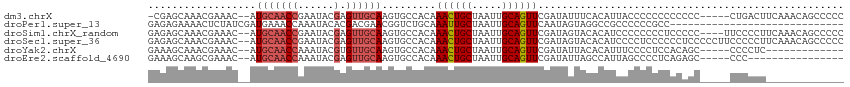
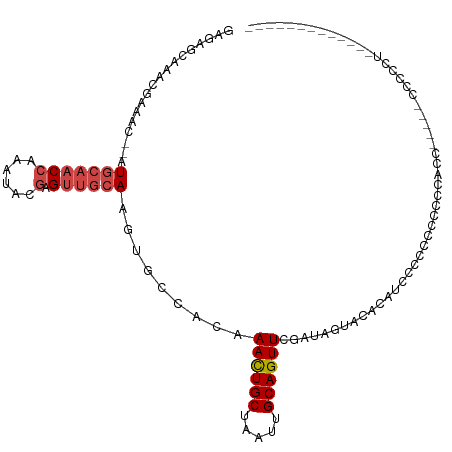
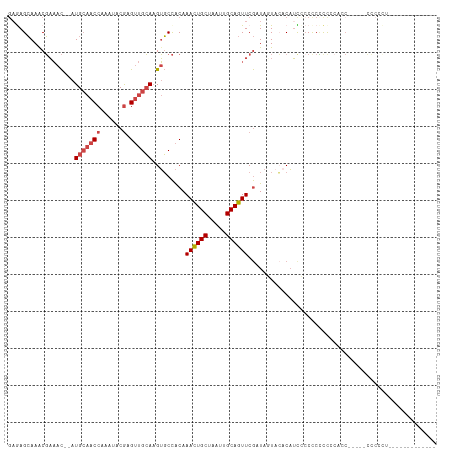
| Location | 11,560,368 – 11,560,459 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.41334 |
| G+C content | 0.42775 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11560368 91 + 22422827 AUGUGAAAUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAUGUUUCGUUUGCUCG-UUUCAAACGCAACGCA-------- ..(((...((.((((((((.....)))))))))).))).((((((.((....)))))))).(((.((((((....-...)))))).)))...-------- ( -28.60, z-score = -2.09, R) >droPer1.super_13 382555 98 - 2293547 CGGCCUACUAUUGAACUGCAAUUAGCAAUUUGCAGACCGUUCGUCGUGUAUUUGGUUUCAUCGAUAG--AGUUUUCUCUCACUUGUCCUGCCACUGUCGG .(((....((((((.((((((........))))))((((.............))))....))))))(--((......))).........)))........ ( -20.22, z-score = -0.05, R) >droSim1.chrX_random 3233159 92 + 5698898 AUGUGUACUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAUGUUUCGUUUGCUCUCUUUCAAACAGCACGCA-------- .(((((.....((((((((.....))))))))..((((.((((((.((....)))))))).))...(((((........))))).)))))))-------- ( -28.80, z-score = -2.74, R) >droSec1.super_36 248476 92 + 449511 AUGUGUACUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAUGUUUCGUUUGCUCUCUUUCAAACACCACGCA-------- .(((((.....((((((((.....)))))))).((.((.((((((.((....)))))))).))...(((((........))))).)))))))-------- ( -27.70, z-score = -2.91, R) >droYak2.chrX 6645427 97 - 21770863 AUGUGUAAUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACACGUAUUUGGUUGCAUGUUUCGUUUGCUUUCUUUCAAAUACAACACAACGCA--- .(((((......(((((((.....)))))))(((((....)).....((((((((..(((.........)))......))))))))..))).)))))--- ( -25.40, z-score = -1.36, R) >consensus AUGUGUACUAUCGAACUGCAAUUAGCAGUUUGUGGCACUUGCAACUCGUAUUCGGUUGCAUGUUUCGUUUGCUCUCUUUCAAACACCACGCA________ .(((((.....((((((((.....)))))))).((.((.((((((.((....)))))))).)).))(((((........)))))...)))))........ (-19.12 = -19.60 + 0.48)

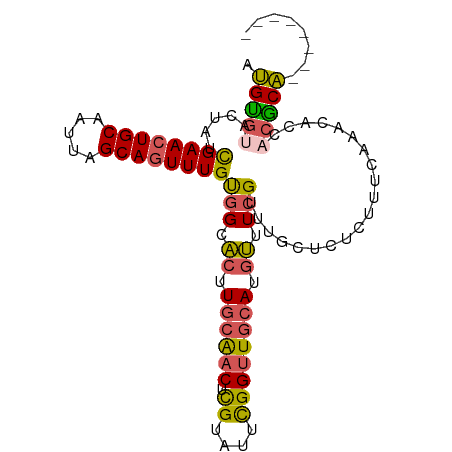
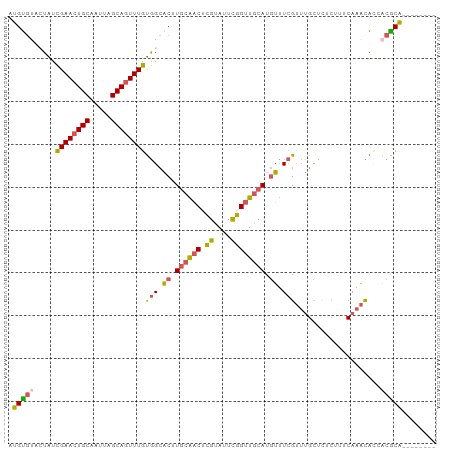
| Location | 11,560,368 – 11,560,459 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.41334 |
| G+C content | 0.42775 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -12.86 |
| Energy contribution | -14.54 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11560368 91 - 22422827 --------UGCGUUGCGUUUGAAA-CGAGCAAACGAAACAUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUUCACAU --------.(((((.((((((...-....)))))).))).((((((((....)).))))))...))..(.((((((.....)))))).)........... ( -25.50, z-score = -1.83, R) >droPer1.super_13 382555 98 + 2293547 CCGACAGUGGCAGGACAAGUGAGAGAAAACU--CUAUCGAUGAAACCAAAUACACGACGAACGGUCUGCAAAUUGCUAAUUGCAGUUCAAUAGUAGGCCG .((...(((...((.....(((.(((....)--)).)))......)).....)))..))..((((((((.((((((.....)))))).....)))))))) ( -22.70, z-score = -1.03, R) >droSim1.chrX_random 3233159 92 - 5698898 --------UGCGUGCUGUUUGAAAGAGAGCAAACGAAACAUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAU --------((.((((((((((........)))))(..((.((((((((....)).)))))).))..).(.((((((.....)))))).)..))))).)). ( -27.30, z-score = -2.50, R) >droSec1.super_36 248476 92 - 449511 --------UGCGUGGUGUUUGAAAGAGAGCAAACGAAACAUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAU --------(((((((((((((........)))))...((.((((((((....)).)))))).))))))).((((((.....)))))).....)))..... ( -28.90, z-score = -3.01, R) >droYak2.chrX 6645427 97 + 21770863 ---UGCGUUGUGUUGUAUUUGAAAGAAAGCAAACGAAACAUGCAACCAAAUACGUGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAUUACACAU ---.....(((((.(((((...............(..((.(((((((......).)))))).))..)...((((((.....)))))).))))).))))). ( -23.20, z-score = -0.64, R) >consensus ________UGCGUGGUGUUUGAAAGAGAGCAAACGAAACAUGCAACCGAAUACGAGUUGCAAGUGCCACAAACUGCUAAUUGCAGUUCGAUAGUACACAU ........((.(((((((.((((..............((.(((((((......).)))))).))........((((.....))))))))))))))).)). (-12.86 = -14.54 + 1.68)

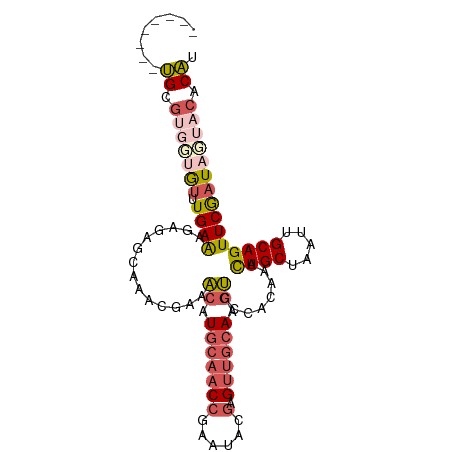
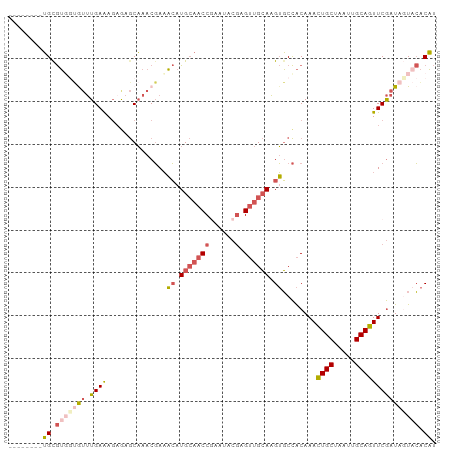
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:35:01 2011