
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,546,216 – 11,546,366 |
| Length | 150 |
| Max. P | 0.831393 |

| Location | 11,546,216 – 11,546,327 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Shannon entropy | 0.32646 |
| G+C content | 0.41267 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -16.49 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11546216 111 - 22422827 GUAGCUCGGUUUUUCCGCAGAACCACAAAUGUAUAUCAAGUACUUUAAUGCUCGAGUGCUCGAGCAUUU-------UCUCUUUGUAAGGUUUUCCUCUCACCCUAUUUGGGAUUUUCC ......(((.....))).(((((((((((.((((.....))))...(((((((((....))))))))).-------....)))))..)))))).......(((.....)))....... ( -26.50, z-score = -2.40, R) >droAna3.scaffold_13335 2795864 113 + 3335858 -UUUGAGAGCAUAUCUGGUUAACCACAA----AUUUCAAGUACCUUAAUGCUUGAAUUCAUUGCAAUUUUGAGUGAUUUCCUUUGUGGGUUUGACGCCCAGCUCUUCCGGAACUUCCC -...((((((......((....))((((----(.((((((((......)))))))).(((((.((....))))))).....)))))((((.....)))).))))))..((.....)). ( -26.80, z-score = -1.37, R) >droEre2.scaffold_4690 15109365 110 + 18748788 GUAGCUCGG-UUUUCCGCAGAACCACAAAUGUAUAUCAAGUACUUUAAUGCUCGAGUGCUCGAGCAUUU-------UCUCUUUGUAAGAUUUUCCUCUCACCCUAUUUGGGAUUUUCC .......((-(((......)))))(((((.((((.....))))...(((((((((....))))))))).-------....)))))...............(((.....)))....... ( -24.90, z-score = -2.02, R) >droYak2.chrX 6632091 110 + 21770863 GUAGCUCGG-UUUUCCGCAGAACCACAAAUGUAUAUCAAGUACUUUAAUGCUCGAGUGCUUGAGCAUUU-------UCUCUUUGUAAGGUUUUCCUCUCACCCUAUUUGGGAUUUUCC .......((-(((......)))))..((((((...((((((((((........))))))))))))))))-------......((..(((....)))..))(((.....)))....... ( -26.70, z-score = -2.27, R) >droSec1.super_36 235356 110 - 449511 GUAGCUCGG-UUUUCCGCAGAACCAAAAAUGUAUAUCAAGUACUUUAAUGCUCGAGUGCUCGAGCAUUU-------UCUCUUUGUAAGGUUUUCCUCUCACCCUAUUUGGGAUUUUCC .......((-(((......)))))......((((.....))))...(((((((((....))))))))).-------......((..(((....)))..))(((.....)))....... ( -25.60, z-score = -1.97, R) >droSim1.chrX 8875667 110 - 17042790 GUAGCUCGG-UUUUCCGCAGAACCACAAAUGUAUAUCAAGUACUUUAAUGCUCGAGUGCUCGAGCAUUU-------UCUCUUUGUAAGGUUUUCCCCUCACCCUAUUUGGGAUUUUCC ......(((-....))).(((((((((((.((((.....))))...(((((((((....))))))))).-------....)))))..)))))).......(((.....)))....... ( -26.50, z-score = -2.25, R) >consensus GUAGCUCGG_UUUUCCGCAGAACCACAAAUGUAUAUCAAGUACUUUAAUGCUCGAGUGCUCGAGCAUUU_______UCUCUUUGUAAGGUUUUCCUCUCACCCUAUUUGGGAUUUUCC ................(.((((((......((((.....))))...(((((((((....)))))))))...................)))))).).....(((.....)))....... (-16.49 = -17.22 + 0.72)

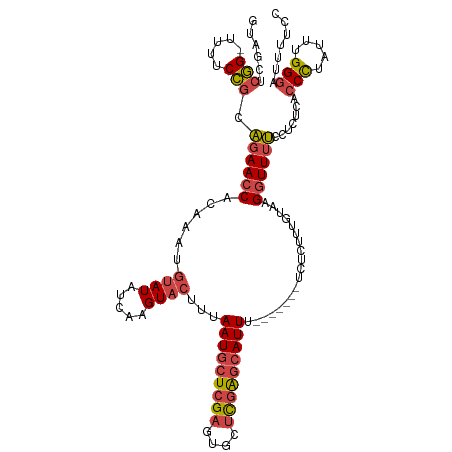

| Location | 11,546,256 – 11,546,366 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Shannon entropy | 0.34215 |
| G+C content | 0.37713 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -14.09 |
| Energy contribution | -15.90 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11546256 110 + 22422827 ---------GAAAAUGCUCGAGCACUCGAGCAUUAAAGUACUUGAUAUACAUUUGUGGUUCUGCGGAAAAACCGAGCUACUUAAUUCCACUGAAUAAUGUCA-UUCCACUUUCAACGACU ---------...(((((((((....)))))))))(((((...((((((......(((((((...((.....)))))))))...((((....)))).))))))-....)))))........ ( -27.00, z-score = -3.05, R) >droAna3.scaffold_13335 2795902 111 - 3335858 GAAAUCACUCAAAAUUGCAAUGAAUUCAAGCAUUAAGGUACUUGAA----AUUUGUGGUUAACCAGAUAU-----GCUCUCAAAGAGAUUUUAGGAAAAUAAUAAUCAUUUUCAAACACU ((((((.((.......(((.....((((((.((....)).))))))----...(.(((....))).)..)-----))......)).))))))..((((((.......))))))....... ( -15.72, z-score = 0.08, R) >droEre2.scaffold_4690 15109405 109 - 18748788 ---------GAAAAUGCUCGAGCACUCGAGCAUUAAAGUACUUGAUAUACAUUUGUGGUUCUGCGGAAAA-CCGAGCUACUUAAUUCCACUGAAUAAUGUCA-UUCCGCUUUCAACGACU ---------...(((((((((....)))))))))(((((...((((((......(((((((...((....-)))))))))...((((....)))).))))))-....)))))........ ( -27.30, z-score = -2.63, R) >droYak2.chrX 6632131 110 - 21770863 ---------GAAAAUGCUCAAGCACUCGAGCAUUAAAGUACUUGAUAUACAUUUGUGGUUCUGCGGAAAA-CCGAGCUACUUAAUUCCACUGAAUAAUGUCAAUUCCGCUUUCAACGACU ---------...(((((((........)))))))(((((..(((((((......(((((((...((....-)))))))))...((((....)))).)))))))....)))))........ ( -22.70, z-score = -1.47, R) >droSec1.super_36 235396 109 + 449511 ---------GAAAAUGCUCGAGCACUCGAGCAUUAAAGUACUUGAUAUACAUUUUUGGUUCUGCGGAAAA-CCGAGCUACUUAAUUCCACUGAAUAAUGUAA-UUCCGCUUUCAACGACU ---------...(((((((((....)))))))))..(((..((((....((....)).....((((((..-......(((...((((....))))...))).-))))))..))))..))) ( -23.00, z-score = -1.74, R) >droSim1.chrX 8875707 109 + 17042790 ---------GAAAAUGCUCGAGCACUCGAGCAUUAAAGUACUUGAUAUACAUUUGUGGUUCUGCGGAAAA-CCGAGCUACUUAAUUCCACUGAAUAAUGUCA-UUCCGCUUUCAACGACU ---------...(((((((((....)))))))))(((((...((((((......(((((((...((....-)))))))))...((((....)))).))))))-....)))))........ ( -27.30, z-score = -2.63, R) >consensus _________GAAAAUGCUCGAGCACUCGAGCAUUAAAGUACUUGAUAUACAUUUGUGGUUCUGCGGAAAA_CCGAGCUACUUAAUUCCACUGAAUAAUGUCA_UUCCGCUUUCAACGACU ............(((((((((....)))))))))..(((..((((.........(((((((............)))))))...((((....))))................))))..))) (-14.09 = -15.90 + 1.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:56 2011