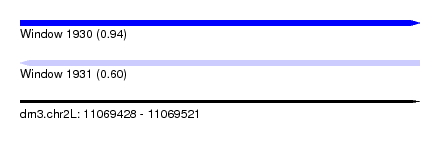
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,069,428 – 11,069,521 |
| Length | 93 |
| Max. P | 0.935601 |

| Location | 11,069,428 – 11,069,521 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 87.72 |
| Shannon entropy | 0.19830 |
| G+C content | 0.57508 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
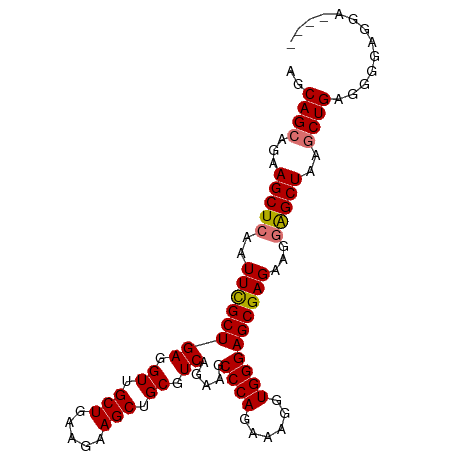
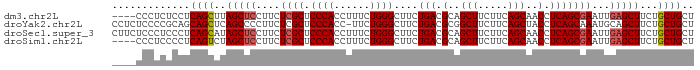
>dm3.chr2L 11069428 93 + 23011544 AGCAGCAGAAGCUCAAUUCGCUGAGGUUGCUGAAGAAGCUGCGUCAGAAGCCCAGAAAGGUGGGAGCGAGAAGGAGCUAAGCUGAGGAGAGGG---- ..((((...(((((..((((((((.((.(((.....))).)).)).....((((......))))))))))...)))))..)))).........---- ( -33.50, z-score = -1.88, R) >droYak2.chr2L 7464792 96 + 22324452 AGCAGCAGAAGCUGCAUUUGCUGAGGUAGCUGAAGAAGCCGCGUCAGAAGCCCAGAA-GGUGGGAGCGAGAAGGGGCUGAGCUGCUGCGGGGAGAGG .(((((((.((((.(.((((((((.(..(((.....)))..).)).....((((...-..))))))))))..).))))...)))))))......... ( -39.00, z-score = -2.09, R) >droSec1.super_3 6470908 97 + 7220098 AGCAGCAGAAGCUCAAUUCGCUGAGGUUGCUGAAGAAGCUGCGUCAGAAGCCCAGAAAGGUGGGAGCGAGAAGGAGCUAUGCUGAGGGAGGGAGAAG ..(((((..(((((..((((((((.((.(((.....))).)).)).....((((......))))))))))...))))).)))))............. ( -36.10, z-score = -2.72, R) >droSim1.chr2L 10867677 93 + 22036055 AGCAGCAGAAGCUCAAUUCGCUGAGGUUGCUGAAGAAGCUGCGUCAGAAGCCCAGAAAGGUGGGAGCGAGAAGGAGCUAGACUGAGGGGAGGG---- ..(..(((.(((((..((((((((.((.(((.....))).)).)).....((((......))))))))))...)))))...)))..)......---- ( -30.60, z-score = -1.04, R) >consensus AGCAGCAGAAGCUCAAUUCGCUGAGGUUGCUGAAGAAGCUGCGUCAGAAGCCCAGAAAGGUGGGAGCGAGAAGGAGCUAAGCUGAGGGGAGGA____ ..((((...(((((..((((((((.((.(((.....))).)).)).....((((......))))))))))...)))))..))))............. (-30.77 = -30.90 + 0.13)


| Location | 11,069,428 – 11,069,521 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.72 |
| Shannon entropy | 0.19830 |
| G+C content | 0.57508 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11069428 93 - 23011544 ----CCCUCUCCUCAGCUUAGCUCCUUCUCGCUCCCACCUUUCUGGGCUUCUGACGCAGCUUCUUCAGCAACCUCAGCGAAUUGAGCUUCUGCUGCU ----.........((((..(((((....((((.((((......))))....(((.(..(((.....)))..).)))))))...)))))...)))).. ( -24.80, z-score = -1.47, R) >droYak2.chr2L 7464792 96 - 22324452 CCUCUCCCCGCAGCAGCUCAGCCCCUUCUCGCUCCCACC-UUCUGGGCUUCUGACGCGGCUUCUUCAGCUACCUCAGCAAAUGCAGCUUCUGCUGCU .........(((((((...(((...........((((..-...))))...((((.(..(((.....)))..).))))........))).))))))). ( -26.60, z-score = -0.92, R) >droSec1.super_3 6470908 97 - 7220098 CUUCUCCCUCCCUCAGCAUAGCUCCUUCUCGCUCCCACCUUUCUGGGCUUCUGACGCAGCUUCUUCAGCAACCUCAGCGAAUUGAGCUUCUGCUGCU .............(((((.(((((....((((.((((......))))....(((.(..(((.....)))..).)))))))...)))))..))))).. ( -25.30, z-score = -1.80, R) >droSim1.chr2L 10867677 93 - 22036055 ----CCCUCCCCUCAGUCUAGCUCCUUCUCGCUCCCACCUUUCUGGGCUUCUGACGCAGCUUCUUCAGCAACCUCAGCGAAUUGAGCUUCUGCUGCU ----.........((((..(((((....((((.((((......))))....(((.(..(((.....)))..).)))))))...)))))...)))).. ( -22.50, z-score = -0.93, R) >consensus ____CCCCCCCCUCAGCUUAGCUCCUUCUCGCUCCCACCUUUCUGGGCUUCUGACGCAGCUUCUUCAGCAACCUCAGCGAAUUGAGCUUCUGCUGCU .............((((..(((((....((((.((((......))))....(((.(..(((.....)))..).)))))))...)))))...)))).. (-21.46 = -22.02 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:30 2011