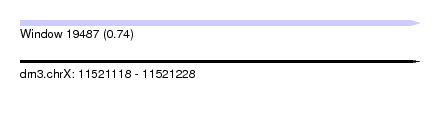
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,521,118 – 11,521,228 |
| Length | 110 |
| Max. P | 0.741779 |

| Location | 11,521,118 – 11,521,228 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 58.12 |
| Shannon entropy | 0.70906 |
| G+C content | 0.29106 |
| Mean single sequence MFE | -17.26 |
| Consensus MFE | -6.00 |
| Energy contribution | -5.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
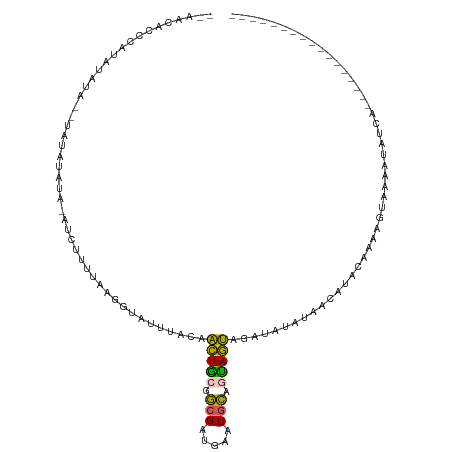
>dm3.chrX 11521118 110 + 22422827 --AACACCCAUAUAUA--UAUAUAUAUAUCUUUUAAGGUAUUUACAACUGCGGCGAAGAAUGCAGUAGUAGAUAUAUAACAUACAAAAUUUAAUUAUCAUUGUGUAACACACUU-- --.((((..(((((((--....)))))))........((((((((.((((((........)))))).))))))))..........................)))).........-- ( -20.20, z-score = -2.02, R) >droSim1.chrX_random 3231334 91 + 5698898 --AACACCCAUAUAUA--UAUAUAU----CUUUUAAGGUAUUUACAACUGCGGCGAUGAAUGCAGUAGUAGAUAUAUAACAUACAAAAGUAACACACUU----------------- --..........((((--(((((((----((....))))))((((.((((((........)))))).))))))))))).....................----------------- ( -15.70, z-score = -1.75, R) >droSec1.super_36 220741 83 + 449511 --AACACCCAUAUAUG--UAUAUAU----CUUUUAAGGUAUUUACAACUGCGGCGAUGAAUGCACUAGUAGAUAUAUAACAUACAAAAGUA------------------------- --..........((((--(((((((----((.(((..((((((((.........).)))))))..))).)))))))).)))))........------------------------- ( -15.50, z-score = -1.37, R) >droYak2.chrX 6617137 116 - 21770863 AAUACACUCAUAUAUAAAUGUAUAUAUAUCUUUGGAGGUAUUCACAACUGCGGCGAUGAGUGCAGCAGUAGUAAUAAAACAUAAAAAAUUAAAUUAUCAAUGCUUAACACACUUAU (((((.((((((((((......))))))).....)))))))).((.(((((.(((.....))).))))).))............................................ ( -17.20, z-score = -0.23, R) >droAna3.scaffold_13335 2777960 97 - 3335858 AAAACUAUGGAAUAUUU-GAUGAGAAAAUGUGAAUAGGAAGUGUCAGUUUAUUAGAAUUCUGAGGAAACAGAGAGACU-UACACGGAAAUCAAAAGUUA----------------- ....((((...((((((-.......))))))..))))...((((.(((((.((((....))))(....)....)))))-.))))...............----------------- ( -17.70, z-score = -1.60, R) >consensus __AACACCCAUAUAUA__UAUAUAUA_AUCUUUUAAGGUAUUUACAACUGCGGCGAUGAAUGCAGUAGUAGAUAUAUAACAUACAAAAGUAAAAUAUCA_________________ ..............................................(((((.(((.....))).)))))............................................... ( -6.00 = -5.20 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:53 2011