| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,511,797 – 11,511,914 |
| Length | 117 |
| Max. P | 0.911201 |

| Location | 11,511,797 – 11,511,914 |
|---|---|
| Length | 117 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.23 |
| Shannon entropy | 0.69404 |
| G+C content | 0.58023 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -12.48 |
| Energy contribution | -13.83 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
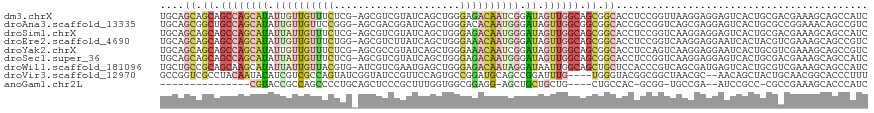
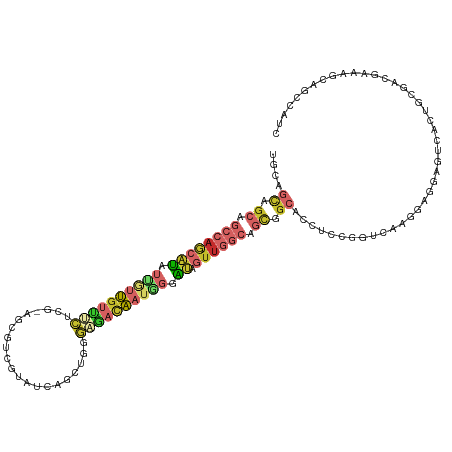
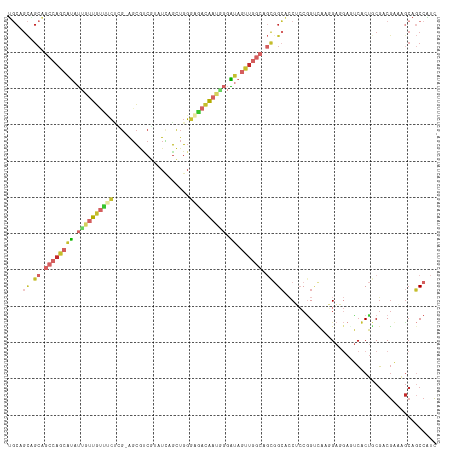
>dm3.chrX 11511797 117 - 22422827 UGCAGCAGCAGCCAGCAUAUUGUUGUUUCUCG-AGCGUCGUAUCAGCUGGGAGACAAUCGGAUAGUUGGCAGCGGCACCUCCGGUUAAGGAGGAGUCACUGCGACGAAAGCAGCCAUC .((((..((.((((((.((((((((((((((.-(((.........)))))))))))))..)))))))))).))(((.(((((......))))).))).))))(.(....))....... ( -48.70, z-score = -3.70, R) >droAna3.scaffold_13335 2767468 117 + 3335858 UGCAGCGGCUGCCAGCAUAUUGUUGUUCCGGG-AGCGACGGAUCAGCUGGGACACAAUGGGAUAGUUGGCGGCGGCACCGCCGGUCAGCGAGGAGUCACUGCGCCGGAAACAGCCGUC ....(((((((.((((.....))))(((((((-((.(((...((.(((((....((((......)))).(((((....))))).)))))))...))).)).).)))))).))))))). ( -49.70, z-score = -1.22, R) >droSim1.chrX 8856200 117 - 17042790 UGCAGCAGCAGCCAGCAUAUUAUUGUUUCUCG-AGCGUCGUAUCAGCUGGGAGACAAUGGGAUAGUUGGCAGCGGCACCUCCGGUCAAGGAGGAGUCACUGCGACGAAAGCAGCCAUC .((((..((.((((((((.((((((((((((.-(((.........))))))))))))))).)).)))))).))(((.(((((......))))).))).))))(.(....))....... ( -49.40, z-score = -4.02, R) >droEre2.scaffold_4690 15085546 117 + 18748788 UGCAGCAGCAGCCAGCAUAUUGUUGUUUCUGG-AGCGUCUUAUCAGCUGGGAAACAAUGGGAUAGUUGGCAGCGGCACCUCCGGUCAAGGAGGAAUCACUACGUCGAAAGCAGCCGUC (((....)))((((((((.(..((((((((.(-.((.........))).))))))))..).)).)))))).(((((.(((((......))))).........(.(....)).))))). ( -44.60, z-score = -2.67, R) >droYak2.chrX 6608018 117 + 21770863 UGCAGCAGCAGCCAGCAUAUUGUUGUUUCUCG-AGCGCCGUAUCAGCUGGGAAACAAUCGGAUAGUUGGCAGCGGCACCUCCAGUCAAGGAGGAAUCACUGCGUCGAAAGCAGCCGUC (((....)))((((((.((((((((((((((.-(((.........)))))))))))))..)))))))))).(((((.(((((......)))))......(((.......)))))))). ( -47.50, z-score = -3.45, R) >droSec1.super_36 211479 117 - 449511 UGCAGCAGCAGCCAGCAUAUUAUUGUUUCUCG-AGCGUCGUAUCAGCUGGGAGACAAUGGGAUAGUUGGCAGCGGCACCUCCGGUCAAGGAGGAGUCACUGCGACGAAAGCAGCCAUC .((((..((.((((((((.((((((((((((.-(((.........))))))))))))))).)).)))))).))(((.(((((......))))).))).))))(.(....))....... ( -49.40, z-score = -4.02, R) >droWil1.scaffold_181096 9614299 117 - 12416693 UGCUGCCGCAGCAAGCAUAUUAUUGUUACGUG-AUCGUCGAAUGAGCUGGGAGACAAUAGGAUAAUUGGCAGCUGCUCCACCCGUCAGCGAUGAGUCACUGCGUCGAAAGCAGCCAUC .(((((((..(((((((......))))..(((-(((((((..(((..((((...((((......))))((....))....))))))).)))))).)))))))..))...))))).... ( -33.00, z-score = 0.62, R) >droVir3.scaffold_12970 1556080 112 + 11907090 GCCGGUCGCCUACAAUACAUCGUCGCCAGUAUCGGUAUCCGUUCCAGUGCCGGAUGCAGCCGGAUUUG----UGGGUACGGCGGCUAACGC--AACAGCUACUGCAACGGCACCCUUU (((((((((((((..((((...(((.(.((((((((((........))))).))))).).)))...))----)).))).))))))....((--(........)))..))))....... ( -38.50, z-score = -0.27, R) >anoGam1.chr2L 24429371 93 + 48795086 ---------------CGUACCGCCAGCCCCUGCAGCUCCCGCUUUGGUGGCGGAGG-AGCUGCUGCUG----CUGCCAC-GCGG-UGCCGA--AUCCGCC-CGCCGAAAGCACCCAUC ---------------.(((((((((((..(.((((((((((((.....))))..))-)))))).)..)----)))....-))))-)))...--.......-.(.(....))....... ( -38.60, z-score = -1.17, R) >consensus UGCAGCAGCAGCCAGCAUAUUGUUGUUUCUCG_AGCGUCGUAUCAGCUGGGAGACAAUGGGAUAGUUGGCAGCGGCACCUCCGGUCAAGGAGGAGUCACUGCGACGAAAGCAGCCAUC ....((.((.((((((((.((((((((((.....................)))))))))).)).)))))).)).)).......................................... (-12.48 = -13.83 + 1.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:50 2011