
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,503,695 – 11,503,785 |
| Length | 90 |
| Max. P | 0.566706 |

| Location | 11,503,695 – 11,503,785 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.24 |
| Shannon entropy | 0.50524 |
| G+C content | 0.50947 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -14.70 |
| Energy contribution | -13.60 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11503695 90 - 22422827 -----------------------UCAACCAAACAA-UACACAACAUG--CACACUACGGCAA--UAGUCGACGGCCAAGCCAGGUGUACUGCCAUGGAGUGGCCUAAAGGUUUUUCCG -----------------------..((((......-...........--.......((((..--..))))..(((((..((((((.....))).)))..)))))....))))...... ( -26.20, z-score = -1.99, R) >droSim1.chrX 8848210 90 - 17042790 -----------------------UCAACCAAACAA-UACACAAUCUG--CACACGACGUCAA--UAGUCGACGGCCAAGUCAGGUGUACUGCCAUGGAGUGGCCUAAAGGUUUUUCCG -----------------------..((((......-...........--....((((.....--..))))..(((((..((((((.....))).)))..)))))....))))...... ( -24.60, z-score = -1.75, R) >droSec1.super_36 202934 90 - 449511 -----------------------UUAACCAACCAA-GACGCGAUCAG--CCCACGGCGUCAA--UAGUCGACGGCCAAGUCAGGUGUACUGCCAUGGAGUGGCCUAAAGGUUUUUCCG -----------------------......((((..-(((((......--......)))))..--........(((((..((((((.....))).)))..)))))....))))...... ( -26.90, z-score = -1.22, R) >droYak2.chrX 6599218 91 + 21770863 -----------------------UCAACCAAUUAAUUACACAACCCG--CACACGAUGACAA--UAGUCGACGGCCAUGCCGCGUGUACUGCCAUGGAGUGGUCUAAAGGUUUUUCCA -----------------------..((((..................--.(((((.((((..--..)))).((((...)))))))))...(((((...))))).....))))...... ( -19.40, z-score = 0.30, R) >droEre2.scaffold_4690 15076838 92 + 18748788 -----------------------UCAACCAAUCAA-UACACAACCCGACAACACGAUGACAA--UAGUCGACGGCCAAGCCGCGUGUACUGCCAUGGAGUGGUCUAAAGGUUUUUCCA -----------------------..((((......-..............(((((.((((..--..)))).((((...)))))))))...(((((...))))).....))))...... ( -19.40, z-score = 0.06, R) >droAna3.scaffold_13117 2210637 118 - 5790199 UCCACCUCAAUCUCCACAACAAACCAACCAACCAACCGAUCGAUCAUCAAUUCCAACCGAAACGCAGUCGCCAGUCGAGCUGCGAGCGCUCGUCAACAGUGGCCUACGGGUGUUUCCC ..(((((......((((....................(((.....))).........(((..(((..((((.((.....))))))))).)))......)))).....)))))...... ( -22.00, z-score = -0.02, R) >consensus _______________________UCAACCAAACAA_UACACAACCAG__CACACGACGACAA__UAGUCGACGGCCAAGCCACGUGUACUGCCAUGGAGUGGCCUAAAGGUUUUUCCG ....................................(((((............((((.........))))..(((...)))..))))).......((((..(((....)))..)))). (-14.70 = -13.60 + -1.10)

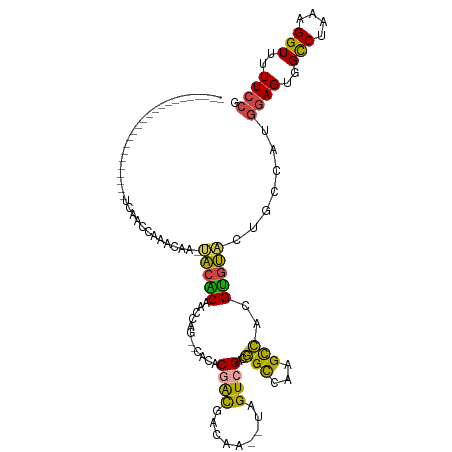
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:49 2011