
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,494,958 – 11,495,072 |
| Length | 114 |
| Max. P | 0.948750 |

| Location | 11,494,958 – 11,495,072 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.43035 |
| G+C content | 0.52415 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -21.80 |
| Energy contribution | -25.08 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11494958 114 - 22422827 AGCAUCUGAUCGUGUCGGUUUUCGCCCGCCGGCCCAUUGAAGGGUUAAA-GGUGGGG---UUUUCUUGGGAGAGUGCGAAGAGUGAAGAGCGCCGCCAUAGCGGAUCGAUGCUCAGAU ((((((.((((((.(((.((((((((((((.((((......))))....-)))))..---(((((....))))).))))))).)))...))..(((....)))))))))))))..... ( -44.90, z-score = -1.95, R) >droEre2.scaffold_4690 15068121 98 + 18748788 AGCAUCUGAUUGUGUCACAUUUCG-CCGCCCGCCCAUUGAAGGGUUAA---CGGCGGGGUGCGUCUUGUGGGAGUGCGAGGAGCGAAGA---UCGAUACA-CAGAU------------ ...(((((..((((((..((..((-(((...((((......))))...---)))))..))((..(((((......)))))..)).....---..))))))-)))))------------ ( -39.10, z-score = -2.24, R) >droYak2.chrX 11087046 101 + 21770863 AGCUUUUGAGUGUGUCACAUUUCG-CCGCCCGCCCAUUGAUGGGUUAAAUUGCAUGGGGUGCAUCUUAUGGGAGUGUGAAGAGCGAAGA---UCGAUACA-CAGAU------------ .........(((((((....((((-((.((((((((....)))))(((..(((((...)))))..))).))).).)))))((.......---))))))))-)....------------ ( -30.70, z-score = -0.87, R) >droSec1.super_36 194426 117 - 449511 AGCAUCUGAUUGUGUCGCUUUUCGCCCGCCGGCCCAUUGAAGGGUUAAA-GGUGGGGUUUUUUUUUUGGUGGAGUGCGAAGAGAGAAGAGCGACGACAUAGCAAAUCGAUGCUCAGAU ((((((.((((.(((((((((((.((((((.((((......))))....-)))))).....((((((.((.....)).)))))))))))))))))........))))))))))..... ( -46.90, z-score = -3.79, R) >droSim1.chrX 8839625 116 - 17042790 AGCAUCUGAUUGUGUCGCUUUUCGCCCGCCGGCCCAUUGAAGGGUUAAA-GGUGGGG-UUUUUUUUUGGUGGAGUGCGAAGAGAGAAGAGCGCCGACAUAGCAGAUCGAUGCUCAGAU ...((((((.(((((((((((((.((((((.((((......))))....-)))))).-...((((((.((.....)).)))))))))))))...))))))(((......))))))))) ( -43.90, z-score = -2.51, R) >consensus AGCAUCUGAUUGUGUCGCUUUUCGCCCGCCGGCCCAUUGAAGGGUUAAA_GGUGGGG_GUUUUUCUUGGGGGAGUGCGAAGAGAGAAGAGCGCCGACAUAGCAGAUCGAUGCUCAGAU ...(((((.((((((((.(((((.(((((((((((......)))))....))))))....((((((((........)))))))))))))....)))))))))))))............ (-21.80 = -25.08 + 3.28)

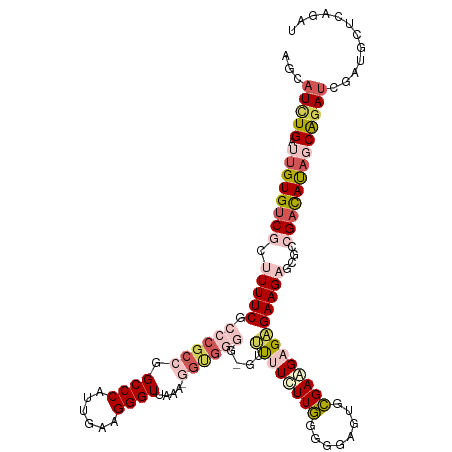
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:47 2011