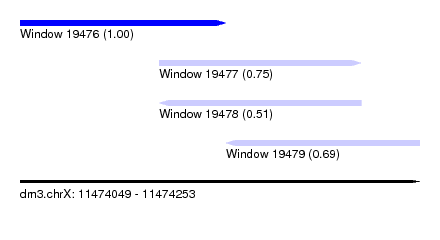
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,474,049 – 11,474,253 |
| Length | 204 |
| Max. P | 0.999247 |

| Location | 11,474,049 – 11,474,154 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.06 |
| Shannon entropy | 0.52333 |
| G+C content | 0.29412 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -12.38 |
| Energy contribution | -11.92 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11474049 105 + 22422827 -----AAGUCUUAAAAGACGUGUAAAAUGUUGCAAAUUAAGCAAAUAUAUA-UGCAUAUAUGGGUAACGUUUUACGCGCCUUAACCAGUCAAAAUACAAAAUAAA--UUGGUA-- -----.........(((.(((((((((((((((..((((.(((........-))).)).))..))))))))))))))).))).((((((...............)--))))).-- ( -26.96, z-score = -3.59, R) >droSim1.chrU 12343312 105 + 15797150 -----AAGUCUUAAAAGACGUGUAAAAUGUUGCAAAUUAAGCAAAUAUAUA-UGCAUAUAUGGGUAAUGUUUUACGCGCCUUAACCAGUCAAAAUACAAAAUAAA--UUGGUA-- -----.........(((.(((((((((((((((..((((.(((........-))).)).))..))))))))))))))).))).((((((...............)--))))).-- ( -24.46, z-score = -2.71, R) >droSec1.super_36 187210 105 + 449511 -----AAGUCUUAAAAGACGUGUAAAAUGUUGCAAAUUGAGCAAAUAUAUA-UGCAUAUAUGGGUAAUGUUUUACGCGCCUUAACCAGUCAAAAUACUAAAUAAA--UUGGUA-- -----.........(((.(((((((((((((((..((...(((........-)))....))..))))))))))))))).))).((((((...............)--))))).-- ( -24.46, z-score = -2.42, R) >droYak2.chrX 11080752 105 - 21770863 -----AAGUCUUAAAAGACGUGUAAAAUGUUGCAAAUUAAGCAAAUAUAUA-UGCAUAUAUGGGUAACGUUUUACGCGCCUUAACCAGUCAAAACACUAAAUAAA--UUGGCA-- -----..((((....))))((((((((((((((..((((.(((........-))).)).))..))))))))))))))(((......(((......))).......--..))).-- ( -26.66, z-score = -3.23, R) >droEre2.scaffold_4690 15060785 103 - 18748788 -----AAGUCUUAAAAGACGUGUAAAAUGUUGCAAAUUAAGCAAAUAUAUA-UGCAUACAUGGGUAACGUUUUACGCGCCUUAACCAGUCAAAAUACUAA--AAA--UGUAUA-- -----..(.((...(((.(((((((((((((((..((((.(((........-))).)).))..))))))))))))))).)))....)).)...((((...--...--.)))).-- ( -24.90, z-score = -3.17, R) >droAna3.scaffold_13417 6463227 105 - 6960332 -----AAGUCUUGAAAGGCAUGCAAAAUGUUGCAAAA-AUAUAUACAUAUAGUAUAUAAAAUGGUAAUGUUUUACGCGCCUUAACCAGCAACCAAACAAAAGUAU---UGAUUU- -----.........(((((.((.((((((((((....-.(((((((.....))))))).....)))))))))).)).))))).......................---......- ( -21.10, z-score = -1.54, R) >dp4.chrXL_group1a 2821804 99 - 9151740 ---CAAAAGUCAAAAAGGCACGUAAAAUGUUGCAAA-------AAAAUGCA-U---UGCAAUGGUAACGUUUUACGAGCCUUAAAAAAUCUCAAAACAAAAAUAUGUUUGAUA-- ---.....(((...(((((.(((((((((((((...-------....(((.-.---.)))...))))))))))))).)))))...........(((((......)))))))).-- ( -26.40, z-score = -4.80, R) >droPer1.super_25 996521 99 + 1448063 ---CAAAAGUCAAAAAGGCACGUAAAAUGUUGCAAA-------AAAAUGCA-U---UGCAAUGGUAACGUUUUACGAGCCUUAAAAAAUCUCAAAACAAAAUUAUGUUUGAUA-- ---.....(((...(((((.(((((((((((((...-------....(((.-.---.)))...))))))))))))).)))))...........(((((......)))))))).-- ( -26.40, z-score = -4.63, R) >droWil1.scaffold_180777 4703622 102 - 4753960 UUAAGACCUAUAAAAAGGCAUGUGAAAUGUUGCAAAA-----AAAAAAAAA------AAAAGGGUAACGUUUUACGCGCCUUAACAAAUC--CAUCUAAAUUUAUAAUUUGUCUA ...((((.(((((((((((.(((((((((((((....-----.........------......))))))))))))).)))))........--........))))))....)))). ( -22.20, z-score = -3.54, R) >consensus _____AAGUCUUAAAAGACGUGUAAAAUGUUGCAAAUUAAGCAAAUAUAUA_UGCAUAUAUGGGUAACGUUUUACGCGCCUUAACCAGUCAAAAUACAAAAUAAA__UUGGUA__ ..............(((.(.(((((((((((((.......((...........))........))))))))))))).).)))................................. (-12.38 = -11.92 + -0.46)


| Location | 11,474,120 – 11,474,223 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Shannon entropy | 0.50680 |
| G+C content | 0.29150 |
| Mean single sequence MFE | -17.16 |
| Consensus MFE | -6.16 |
| Energy contribution | -5.81 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11474120 103 + 22422827 GCCUUAACCAGUCAAAAUACAAAAUAAA--UUGGUAAAUUUCAUAUAACUAGUGAAAUGUUAUACGAAAC-UUAACAAUUGCCAAAUAAUACAGAUCGAUUUAGAG------ ............................--((((((((((((((.......)))))))((((........-.))))..))))))).....................------ ( -12.00, z-score = -0.84, R) >droSim1.chrU 12343383 103 + 15797150 GCCUUAACCAGUCAAAAUACAAAAUAAA--UUGGUAAAUUUCAUAUAACUAGUGAAAUGUUAUACGAAAC-UUAACAAUUGCCAAAUAAUACAGAUCUAUUUAGAG------ ............................--((((((((((((((.......)))))))((((........-.))))..))))))).....................------ ( -12.00, z-score = -0.89, R) >droSec1.super_36 187281 103 + 449511 GCCUUAACCAGUCAAAAUACUAAAUAAA--UUGGUAAAUUUCAUAUAACUAGUGAAAUGUUAUACGAAAC-UUAACAAUUGCCAAAUAAUACAGAUCUAUUUAGAG------ ...................(((((((..--((((((((((((((.......)))))))((((........-.))))..)))))))............)))))))..------ ( -13.34, z-score = -1.30, R) >droYak2.chrX 11080823 103 - 21770863 GCCUUAACCAGUCAAAACACUAAAUAAA--UUGGCAAAUUUCAUAUAACUAGUGAAAUGUUAUACGAAAC-UUGCCAAUUGUCAAAUAAUCCAGAUCUAUUUAGAG------ ...................(((((((((--(((((((.((((.((((((.........)))))).)))).-)))))))))(((..........))).)))))))..------ ( -22.60, z-score = -4.53, R) >droEre2.scaffold_4690 15060856 101 - 18748788 GCCUUAACCAGUCAAAAUACUAA--AAA--UGUAUAAAUUUCAUAUAACUAGUGAAAUGUUGUACGAAAC-UUAGCAAUUGUCAAGUUAUUCAGAUCUAUUUAGAG------ ...................((((--(..--((((((((((((((.......))))))).))))))).(((-((..(....)..)))))...........)))))..------ ( -15.90, z-score = -1.44, R) >droAna3.scaffold_13417 6463298 110 - 6960332 GCCUUAACCAGCAACCAAACAAAAGUAU--UGAUUUUUUUUCGUAUGACGCAUGCAACGUUAUACGAAACAUUCUCAAUAAACAAAAAACACAAAAGCGUCUUCAAUUCGAG ((........)).............(((--(((.....((((((((((((.......)))))))))))).....))))))................................ ( -18.20, z-score = -2.41, R) >dp4.chrXL_group1a 2821867 98 - 9151740 GCCUUAAAAAAUCUCAAAACAAAAAUAUGUUUGAUAAAUUUCGUAUGACUCAUGAAAUGUGAUACGAAAU-UUGUUGAUUAUCG-------CCGACUUGUUCAGAG------ ............(((..(((((....(((.(..(((((((((((((.((.........)).)))))))))-))))..).)))..-------.....)))))..)))------ ( -21.60, z-score = -3.16, R) >droPer1.super_25 996584 98 + 1448063 GCCUUAAAAAAUCUCAAAACAAAAUUAUGUUUGAUAAAUUUCGUAUGACUCAUGAAAUGUGAUACGAAAU-UUGUUGAUUAUCG-------CCGACUUGUUCAGAG------ ............(((..(((((....(((.(..(((((((((((((.((.........)).)))))))))-))))..).)))..-------.....)))))..)))------ ( -21.60, z-score = -3.02, R) >consensus GCCUUAACCAGUCAAAAUACAAAAUAAA__UUGGUAAAUUUCAUAUAACUAGUGAAAUGUUAUACGAAAC_UUAACAAUUGUCAAAUAAUACAGAUCUAUUUAGAG______ ..............................(((((((.((((.((((((.........)))))).)))).........)))))))........................... ( -6.16 = -5.81 + -0.34)


| Location | 11,474,120 – 11,474,223 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Shannon entropy | 0.50680 |
| G+C content | 0.29150 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -6.48 |
| Energy contribution | -6.34 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11474120 103 - 22422827 ------CUCUAAAUCGAUCUGUAUUAUUUGGCAAUUGUUAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUUUACCAA--UUUAUUUUGUAUUUUGACUGGUUAAGGC ------.(((.(((((.((.((((........(((((.(((-((((((((((((.........))))))))))))))).)))--))......))))...)).))))).))). ( -22.44, z-score = -3.01, R) >droSim1.chrU 12343383 103 - 15797150 ------CUCUAAAUAGAUCUGUAUUAUUUGGCAAUUGUUAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUUUACCAA--UUUAUUUUGUAUUUUGACUGGUUAAGGC ------(..(((.(((.((.((((........(((((.(((-((((((((((((.........))))))))))))))).)))--))......))))...))))).)))..). ( -20.14, z-score = -1.95, R) >droSec1.super_36 187281 103 - 449511 ------CUCUAAAUAGAUCUGUAUUAUUUGGCAAUUGUUAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUUUACCAA--UUUAUUUAGUAUUUUGACUGGUUAAGGC ------(..(((.(((.((.((((((..((..(((((.(((-((((((((((((.........))))))))))))))).)))--))))..))))))...))))).)))..). ( -23.20, z-score = -2.99, R) >droYak2.chrX 11080823 103 + 21770863 ------CUCUAAAUAGAUCUGGAUUAUUUGACAAUUGGCAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUUUGCCAA--UUUAUUUAGUGUUUUGACUGGUUAAGGC ------..(((((((((..((..........)).(((((((-((((((((((((.........)))))))))))))))))))--))))))))).(((((((....))))))) ( -31.10, z-score = -4.97, R) >droEre2.scaffold_4690 15060856 101 + 18748788 ------CUCUAAAUAGAUCUGAAUAACUUGACAAUUGCUAA-GUUUCGUACAACAUUUCACUAGUUAUAUGAAAUUUAUACA--UUU--UUAGUAUUUUGACUGGUUAAGGC ------....................((((((......(((-((((((((.(((.........))).)))))))))))....--...--.((((......)))))))))).. ( -16.60, z-score = -0.78, R) >droAna3.scaffold_13417 6463298 110 + 6960332 CUCGAAUUGAAGACGCUUUUGUGUUUUUUGUUUAUUGAGAAUGUUUCGUAUAACGUUGCAUGCGUCAUACGAAAAAAAAUCA--AUACUUUUGUUUGGUUGCUGGUUAAGGC .((((((.((((((((....))))........((((((.....((((((((.((((.....)))).)))))))).....)))--))))))).))))))..(((......))) ( -27.80, z-score = -2.08, R) >dp4.chrXL_group1a 2821867 98 + 9151740 ------CUCUGAACAAGUCGG-------CGAUAAUCAACAA-AUUUCGUAUCACAUUUCAUGAGUCAUACGAAAUUUAUCAAACAUAUUUUUGUUUUGAGAUUUUUUAAGGC ------..........(((..-------....((((...((-(((((((((.((.........)).))))))))))).(((((((......)).)))))))))......))) ( -17.60, z-score = -1.45, R) >droPer1.super_25 996584 98 - 1448063 ------CUCUGAACAAGUCGG-------CGAUAAUCAACAA-AUUUCGUAUCACAUUUCAUGAGUCAUACGAAAUUUAUCAAACAUAAUUUUGUUUUGAGAUUUUUUAAGGC ------..........(((..-------....((((...((-(((((((((.((.........)).))))))))))).(((((((......)).)))))))))......))) ( -17.60, z-score = -1.44, R) >consensus ______CUCUAAAUAGAUCUGUAUUAUUUGACAAUUGAUAA_GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUUUACCAA__UUUAUUUUGUAUUUUGACUGGUUAAGGC ..........................................(((((((((.((.........)).)))))))))..................................... ( -6.48 = -6.34 + -0.14)


| Location | 11,474,154 – 11,474,253 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.05 |
| Shannon entropy | 0.48994 |
| G+C content | 0.35903 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -7.62 |
| Energy contribution | -7.75 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.690181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11474154 99 - 22422827 ---GAGAGAGAUCGCCUCUCUAUUGUCAUCUCGCUCUAAAUCGA------UCUGUAUUAUUUGGCAAUUGUUAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUU ---.((((((.....))))))(((((((................------...........))))))).....(-((((((((((((.........))))))))))))) ( -21.83, z-score = -2.11, R) >droSim1.chrU 12343417 99 - 15797150 ---GAGAGAGAUCGCCUCUCUAUGGCCAUCUCGCUCUAAAUAGA------UCUGUAUUAUUUGGCAAUUGUUAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUU ---(((.(((((.(((.......))).))))).)))........------..........(((((....)))))-((((((((((((.........)))))))))))). ( -27.30, z-score = -3.53, R) >droSec1.super_36 187315 99 - 449511 ---GAGAGAGAUCGCCUCUCUAUUGCCAUCUCGCUCUAAAUAGA------UCUGUAUUAUUUGGCAAUUGUUAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUU ---.((((((.....))))))(((((((....(((((....)))------...))......))))))).....(-((((((((((((.........))))))))))))) ( -25.00, z-score = -3.13, R) >droYak2.chrX 11080857 99 + 21770863 ---GAGGGAGAUCACCUCUCUAUUGCCAUCUUGCUCUAAAUAGA------UCUGGAUUAUUUGACAAUUGGCAA-GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUU ---.((((((.....)))))).((((((..(((.((.((((((.------......))))))))))).))))))-((((((((((((.........)))))))))))). ( -26.90, z-score = -3.19, R) >droEre2.scaffold_4690 15060888 99 + 18748788 ---GAGAGAGAUCACCUCUCUAUUGCCAUCCUGCUCUAAAUAGA------UCUGAAUAACUUGACAAUUGCUAA-GUUUCGUACAACAUUUCACUAGUUAUAUGAAAUU ---.((((((.....)))))).......................------.......................(-((((((((.(((.........))).))))))))) ( -14.20, z-score = 0.03, R) >droAna3.scaffold_13417 6463332 106 + 6960332 ---GAAAGAGAUCGCCAAUGUCUAUCUGUCUCGCUCGAAUUGAAGACGCUUUUGUGUUUUUUGUUUAUUGAGAAUGUUUCGUAUAACGUUGCAUGCGUCAUACGAAAAA ---((.(((.((.....)).))).))..(((((...((((.((((((((....)))))))).))))..)))))...((((((((.((((.....)))).)))))))).. ( -28.40, z-score = -1.90, R) >dp4.chrXL_group1a 2821903 94 + 9151740 UCUGAAAGAGAUCGCCUCUAAGC-UCCAUCCUGCUCUGAACAAG------UCGG-------CGAUAAUCAACAA-AUUUCGUAUCACAUUUCAUGAGUCAUACGAAAUU .......((.((((((((..(((-........)))..)).....------..))-------))))..))....(-(((((((((.((.........)).)))))))))) ( -20.81, z-score = -1.96, R) >droPer1.super_25 996620 94 - 1448063 UCUGAAAGAUAUCGCCUCUAAGC-UCCAUCCUGCUCUGAACAAG------UCGG-------CGAUAAUCAACAA-AUUUCGUAUCACAUUUCAUGAGUCAUACGAAAUU .......(((((((((((..(((-........)))..)).....------..))-------)))).)))....(-(((((((((.((.........)).)))))))))) ( -22.61, z-score = -3.17, R) >consensus ___GAGAGAGAUCGCCUCUCUAUUGCCAUCUCGCUCUAAAUAGA______UCUGUAUUAUUUGACAAUUGAUAA_GUUUCGUAUAACAUUUCACUAGUUAUAUGAAAUU ......((((.....))))........................................................(((((((((.((.........)).))))))))). ( -7.62 = -7.75 + 0.13)

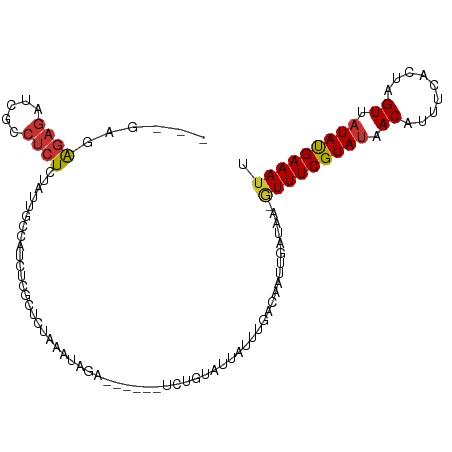

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:46 2011