| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,461,253 – 11,461,373 |
| Length | 120 |
| Max. P | 0.765969 |

| Location | 11,461,253 – 11,461,373 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.68 |
| Shannon entropy | 0.32208 |
| G+C content | 0.48667 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -19.74 |
| Energy contribution | -18.43 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
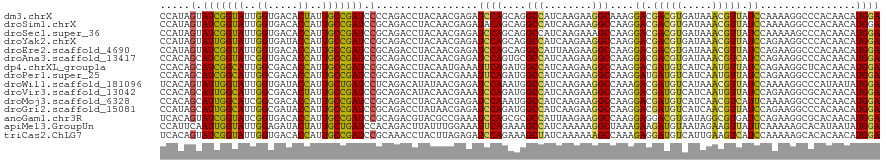
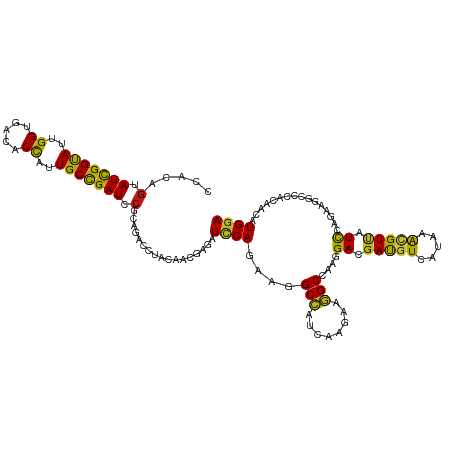
>dm3.chrX 11461253 120 - 22422827 CCAUAGUAUCGGUAUUGGUGACACUAUUGCCGAUCCCCAGACCUACAACGAGAUCCAGCAGGCCAUCAAGAAGGCAAAGGACGACGUGAUAAACGUUAUCCAAAAGGCCCACAACAUGGA ((((.(((((((((.(((.....))).))))))).......(((.................(((........)))...(((..((((.....))))..)))...)))......)))))). ( -31.70, z-score = -1.72, R) >droSim1.chrX 8827622 120 - 17042790 CCAUAGUAUCGGUAUUGGUGACACCAUUGCCGAUCCGCAGACCUACAACGAGAUACAGCAGGCCAUCAAGAAGGCCAAGGACGACGUGAUAAACGUUAUCCAAAAGGCCCACAACAUGGA ((((.(.(((((((.(((.....))).))))))).).....(((................((((........))))..(((..((((.....))))..)))...)))........)))). ( -38.00, z-score = -3.74, R) >droSec1.super_36 174747 120 - 449511 CCAUAGUAUCGGUAUUGGUGACACCAUUGCCGAUCCGCAGACCUACAACGAGAUCCAGCAGGCCAUCAAGAAAGCCAAGGACGACGUGAUAAACGUUAUCCAAAAAGCCCACAACAUGGA ((((.(.(((((((.(((.....))).))))))).)((......................(((..........)))..(((..((((.....))))..))).....)).......)))). ( -32.90, z-score = -2.97, R) >droYak2.chrX 11068290 120 + 21770863 CCAUAGUAUUGGUAUUGGUGAUACCAUUGCCGAUCCGCAGACCUACAACGAGAUCCAGCAGGCCAUCAAGAAGGCCAAGGACGACGUGAUAAACGUUAUCCAGAAGGCCCACAACAUGGA ((((.(.(((((((.(((.....))).))))))).).....(((.(..............((((........))))..(((..((((.....))))..))).).)))........)))). ( -36.20, z-score = -2.85, R) >droEre2.scaffold_4690 15048369 120 + 18748788 CCAUAGUAUCGGUAUUGGUGACACCAUUGCCGAUCCGCAGACCUACAACGAGAUCCAGCAGGCCAUUAAGAAGGCCAAGGACGACGUGAUAAACGUUAUCCAGAAGGCCCACAACAUGGA ((((.(.(((((((.(((.....))).))))))).).....(((.(..............((((........))))..(((..((((.....))))..))).).)))........)))). ( -38.30, z-score = -3.62, R) >droAna3.scaffold_13417 6450361 120 + 6960332 CCACAGCAUCGGUAUCGGUGACACCAUUGCCGAUCCGCAGACCUACAACGAGAUCCAGUGCGCCAUCAAGAAGGCCAAGGACGACGUGAUAAACGUCAUCCAGAAGGCCCACAACAUGGA .....(((((((.(((((..(.....)..))))))))..((.((......)).))..)))).((((......((((..(((.(((((.....))))).)))....))))......)))). ( -37.10, z-score = -2.43, R) >dp4.chrXL_group1a 2807022 120 + 9151740 CCACAGCAUCGGCAUUGGCGACACCAUUGCCGAUCCGCAGACCUACAAUGAAAUUCAGAUGGCCAUCAAGAAGGCCAAGGACGAUGUCAUCAAUGUUAUCCAGAAGGCUCACAACAUGGA (((..(((((((((.(((.....))).)))))))..)).(((((.(.............(((((........))))).(((..((((.....))))..))).).))).))......))). ( -36.50, z-score = -2.23, R) >droPer1.super_25 979079 120 - 1448063 CCACAGCAUCGGCAUUGGCGACACCAUUGCCGAUCCGCAGACCUACAACGAAAUUCAGAUGGCCAUCAAGAAGGCCAAGGAUGAUGUCAUCAAUGUUAUCCAGAAGGCCCACAACAUGGA (((..(((((((((.(((.....))).)))))))..))...(((.(.............(((((........))))).(((((((((.....))))))))).).))).........))). ( -37.80, z-score = -2.67, R) >droWil1.scaffold_181096 11223132 120 + 12416693 UCACAGUAUUGGUAUUGGUGAUACCAUUGCUGAUCCUCAGACAUAUAACGAGAUCCAAAUGGCCAUCAAGAAGGCCAAAGACGAUGUCAUAAACGUUAUCCAAAAGGCCCAUAAUAUGGA ...(((((.(((((((...))))))).)))))..(((.......((((((.........(((((........)))))..(((...))).....)))))).....))).((((...)))). ( -30.00, z-score = -1.55, R) >droVir3.scaffold_13042 1856316 120 + 5191987 CCACAGCAUUGGCAUUGGCGACACUAUUGCCGAUCCGCAGACAUACAACGAAAUCCAGAUGGCCAUCAAGAAGGCCAAGGACGAUGUCAUCAAUGUUAUCCAGAAGGCGCACAACAUGGA ....(((((((((((((((((.....))))))))..)).(((((....((...(((...(((((........))))).))))))))))..))))))).((((....(....)....)))) ( -33.00, z-score = -1.46, R) >droMoj3.scaffold_6328 648379 120 + 4453435 CCACAGCAUUGGCAUCGGCGACACCAUUGCCGAUCCGCAGACCUACAACGAGAUCCAAAUGGCCAUCAAGAAGGCCAAGGACGAUGUCAUCAACGUCAUUCAAAAGGCCCACAACAUGGA (((..((...((.((((((((.....))))))))))))...(((.........(((...(((((........))))).))).(((((.....))))).......))).........))). ( -32.80, z-score = -1.61, R) >droGri2.scaffold_15081 2946367 120 - 4274704 CCAUAGCAUUGGCAUUGGCGAUACCAUUGCCGAUCCGCAGACCUAUAACGAGAUCCAGAUGGCCAUCAAGAAGGCCAAGGACGAUGUCAUCAACGUUAUCCAGAAGGCCCACAACAUGGA ((((.(((((((((.(((.....))).)))))))..))...(((...............(((((........))))).(((..((((.....))))..)))...)))........)))). ( -36.10, z-score = -2.37, R) >anoGam1.chr3R 1320610 120 - 53272125 UCACAGUAUCGGUAUCGGUGACACCAUUGCCGAUCCGCAGACGUACGCCGAAAUCCAGCGCGCCAUUAAGAAGGCCAAGGAGGACGUGAUAGGCGUGAUCCAGAAGGCGCACAACAUGGA .....((..(((.(((((..(.....)..))))))))...))((.((((.........((((((((((....(.((.....)).).)))).))))))........)))).))........ ( -36.63, z-score = -0.75, R) >apiMel3.GroupUn 146906886 120 + 399230636 CCAUUCAAUUGGUAUUGGAGAUACUAUUGCUGAUCCACAGACUUAUUUGGAAAUUCAGAAAGCCAUCAAAAAGGCUAAAGAAGAUGUAAUAGAAGUUAUUCAAAAAGCACAUAAUAUGGA ((((.....((((.(((((....((((((((((((((.(......).))))...)))...((((........)))).........)))))))......)))))...)).))....)))). ( -20.60, z-score = -0.17, R) >triCas2.ChLG7 16085235 120 + 17478683 UCACAGUAUCGGUAUUGGUGACACCAUUGCCGAUCCGCAAACCUACUUAGAGAUCCAGAAAGCUAUCAAAAAAGCCAAAGAGGAUGUCAUUGAAGUCAUCCAAAAAGCACACAACAUGGA .....(.(((((((.(((.....))).))))))).).................((((....(((........)))......(((((.(......).)))))...............)))) ( -26.60, z-score = -1.59, R) >consensus CCACAGUAUCGGUAUUGGUGACACCAUUGCCGAUCCGCAGACCUACAACGAGAUCCAGAAGGCCAUCAAGAAGGCCAAGGACGAUGUCAUAAACGUUAUCCAGAAGGCCCACAACAUGGA .....(.(((((((..((.....))..))))))).).................((((....(((........)))....((.(((((.....))))).))................)))) (-19.74 = -18.43 + -1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:41 2011