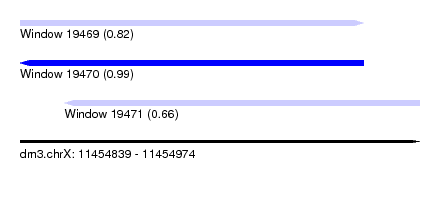
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,454,839 – 11,454,974 |
| Length | 135 |
| Max. P | 0.992376 |

| Location | 11,454,839 – 11,454,955 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 68.41 |
| Shannon entropy | 0.58217 |
| G+C content | 0.43035 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
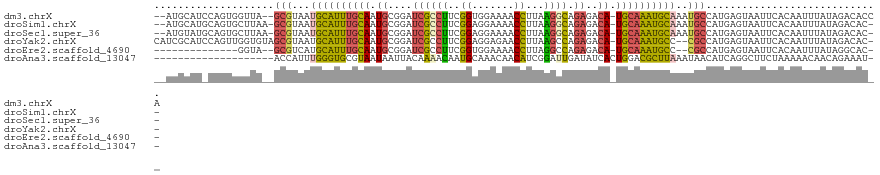
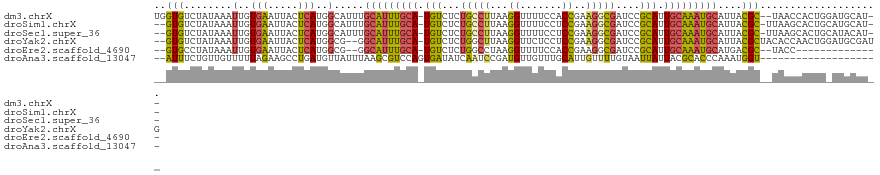
>dm3.chrX 11454839 116 + 22422827 --AUGCAUCCAGUGGUUA--GCGUAAUGCAUUUGCAAUGCGGAUCGCCUUCGGUGGAAAACCUUAAGGCAGAGACA-UGCAAAUGCAAAUGCCAUGAGUAAUUCACAAUUUAUAGACACCA --.........((((...--((...(((((((((((.((((..(((((((.(((.....)))..))))).))....-))))..)))))))).)))..))...))))............... ( -31.90, z-score = -1.12, R) >droSim1.chrX 8821907 115 + 17042790 --AUGCAUGCAGUGCUUAA-GCGUAAUGCAUUUGCAAUGCGGAUCGCCUUCGGAGGAAAACCUUAAGGCAGAGACA-UGCAAAUGCAAAUGCCAUGAGUAAUUCACAAUUUAUAGACAC-- --.....((...((((((.-((((..((((((((((....(..(((((((..((((....))))))))).))..).-)))))))))).))))..))))))...))..............-- ( -32.40, z-score = -1.64, R) >droSec1.super_36 168396 115 + 449511 --AUGUAUGCAGUGCUUAA-GCGUAAUGCAUUUGCAAUGCGGAUCGCCUUCGGAGGAAAACCUUAAGGCAGAGACA-UGCAAAUGCAAAUGCCAUGAGUAAUUCACAAUUUAUAGACAC-- --.(((......((((((.-((((..((((((((((....(..(((((((..((((....))))))))).))..).-)))))))))).))))..))))))....)))............-- ( -32.70, z-score = -1.97, R) >droYak2.chrX 11061967 116 - 21770863 CAUCGCAUCCAGUUGGUGUAGCGUAAUGCAUUUGCAAUGCGGAUCGCCUUCGGAGGAGAACCUUAAGCCAGAGACA-UGCAAAUGCC--CGCCAUGAGUAAUUCACAAUUUAUAGACAC-- ....(((((.....))))).(((....(((((((((..(((...)))((..((..(((...)))...))..))...-))))))))).--))).((((((........))))))......-- ( -24.50, z-score = 0.88, R) >droEre2.scaffold_4690 15042028 100 - 18748788 --------------GGUA--GCGUCAUGCAUUUGCAAUGCGGAUCGCCUUCGGUGGAAAACCUUAGGCCAGAGACA-UGCAAAUGCC--CGCCAUGAGUAAUUCACAAUUUAUAGGCAC-- --------------....--.......(((((((((.(((.....((((..(((.....)))..))))....).))-))))))))).--.(((((((((........)))))).)))..-- ( -30.20, z-score = -1.31, R) >droAna3.scaffold_13047 199930 99 + 1816235 --------------------ACCAUUUGGGUGCGUAAUAAUUACAAAACAAUGCAAACAACAUCGGAUUGAUAUCACUGGACGCUUAAAUAACAUCAGGCUUCUAAAAACAACAGAAAU-- --------------------.(((((((..(((((...............)))))..))).)).))...........((((.((((..........)))).))))..............-- ( -12.06, z-score = -0.17, R) >consensus __AUGCAU_CAGUGGUUA__GCGUAAUGCAUUUGCAAUGCGGAUCGCCUUCGGAGGAAAACCUUAAGGCAGAGACA_UGCAAAUGCAAAUGCCAUGAGUAAUUCACAAUUUAUAGACAC__ ....................(((...((((((((((.((....((((((..((.......))...)))).))..)).))))))))))..)))............................. (-15.98 = -15.65 + -0.33)
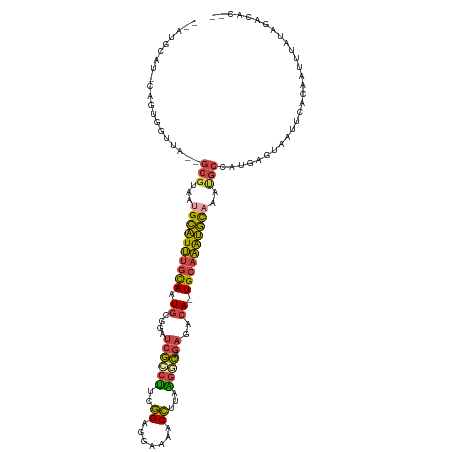
| Location | 11,454,839 – 11,454,955 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 68.41 |
| Shannon entropy | 0.58217 |
| G+C content | 0.43035 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -17.37 |
| Energy contribution | -16.63 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11454839 116 - 22422827 UGGUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCA-UGUCUCUGCCUUAAGGUUUUCCACCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGC--UAACCACUGGAUGCAU-- ..(((((((......((((.....))))(((...((((((((((-(((...((((((..(((.....))).))))))....))).))))))))))....))--)......)))))))..-- ( -35.30, z-score = -2.29, R) >droSim1.chrX 8821907 115 - 17042790 --GUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCA-UGUCUCUGCCUUAAGGUUUUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGC-UUAAGCACUGCAUGCAU-- --(((..((......((((.....))))(((...((((((((((-(((...((((((.(((....)))...))))))....))).))))))))))....))-)))..))).........-- ( -32.80, z-score = -1.80, R) >droSec1.super_36 168396 115 - 449511 --GUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCA-UGUCUCUGCCUUAAGGUUUUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGC-UUAAGCACUGCAUACAU-- --(((..((......((((.....))))(((...((((((((((-(((...((((((.(((....)))...))))))....))).))))))))))....))-)))..))).........-- ( -32.80, z-score = -2.37, R) >droYak2.chrX 11061967 116 + 21770863 --GUGUCUAUAAAUUGUGAAUUACUCAUGGCG--GGCAUUUGCA-UGUCUCUGGCUUAAGGUUCUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGCUACACCAACUGGAUGCGAUG --(((((((......((((.....))))((((--.(((((((((-(((...(.((((..((.......))..)))).)...))).)))))))))....))))........))))))).... ( -34.30, z-score = -1.52, R) >droEre2.scaffold_4690 15042028 100 + 18748788 --GUGCCUAUAAAUUGUGAAUUACUCAUGGCG--GGCAUUUGCA-UGUCUCUGGCCUAAGGUUUUCCACCGAAGGCGAUCCGCAUUGCAAAUGCAUGACGC--UACC-------------- --.............((((.....))))((((--.(((((((((-(((...(.((((..(((.....)))..)))).)...))).)))))))))....)))--)...-------------- ( -31.70, z-score = -1.69, R) >droAna3.scaffold_13047 199930 99 - 1816235 --AUUUCUGUUGUUUUUAGAAGCCUGAUGUUAUUUAAGCGUCCAGUGAUAUCAAUCCGAUGUUGUUUGCAUUGUUUUGUAAUUAUUACGCACCCAAAUGGU-------------------- --..(((((.......)))))((..((((((.....))))))..((((((.(((..((((((.....))))))..)))....))))))))(((.....)))-------------------- ( -21.40, z-score = -2.16, R) >consensus __GUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCA_UGUCUCUGCCUUAAGGUUUUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGC__UAACCACUG_AUGCAU__ ..(((........(..(((.....)))..).....(((((((((.(((...(((((...((.......))..)))))....))).)))))))))....))).................... (-17.37 = -16.63 + -0.74)
| Location | 11,454,854 – 11,454,974 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.41 |
| Shannon entropy | 0.48722 |
| G+C content | 0.41613 |
| Mean single sequence MFE | -33.09 |
| Consensus MFE | -12.64 |
| Energy contribution | -14.73 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.661857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
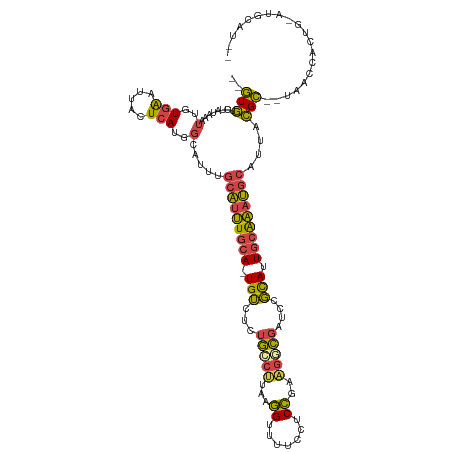
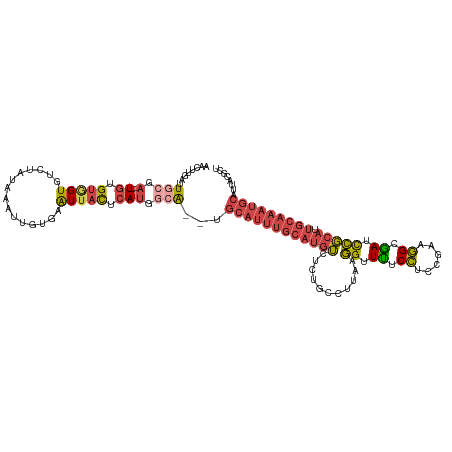
>dm3.chrX 11454854 120 - 22422827 AACUUGAUUGCGAUGUGUGUGGUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCAUGUCUCUGCCUUAAGGUUUUCCACCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGCU .........(((..(((.(((((..(........)...))))).))).(((((((((..(((..(((...(((((..(((.....))).))))))))..))).)))))))))....))). ( -34.70, z-score = -1.55, R) >droSim1.chrX 8821923 118 - 17042790 AACUUGAUUGCGA--UGAGUGGUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCAUGUCUCUGCCUUAAGGUUUUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGCU ........(((.(--((((((((..(........)...))))))))).)))..(((((((((((((...((((((.(((....)))...))))))....))).))))))))))....... ( -40.40, z-score = -3.81, R) >droSec1.super_36 168412 118 - 449511 AACUUCAUUGCGA--UGUGUGGUGUCUAUAAAUUGUGAAUUACUCAUGGCAUUUGCAUUUGCAUGUCUCUGCCUUAAGGUUUUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGCU ........(((.(--((.(((((..(........)...))))).))).)))..(((((((((((((...((((((.(((....)))...))))))....))).))))))))))....... ( -36.20, z-score = -2.75, R) >droYak2.chrX 11061986 116 + 21770863 AACUUGAUUCCGA--UGUGCAGUGUCUAUAAAUUGUGAAUUACUCAUGGCG--GGCAUUUGCAUGUCUCUGGCUUAAGGUUCUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGCU ....(((((((((--(...............)))).)))))).....((((--.((((((((((((...(.((((..((.......))..)))).)...))).)))))))))....)))) ( -30.66, z-score = -0.32, R) >droEre2.scaffold_4690 15042031 116 + 18748788 AACUUGAUUCAUA--UGUGUAGUGCCUAUAAAUUGUGAAUUACUCAUGGCG--GGCAUUUGCAUGUCUCUGGCCUAAGGUUUUCCACCGAAGGCGAUCCGCAUUGCAAAUGCAUGACGCU ....(((((((((--..(((((...)))))...))))))))).....((((--.((((((((((((...(.((((..(((.....)))..)))).)...))).)))))))))....)))) ( -39.40, z-score = -2.83, R) >droAna3.scaffold_13047 199946 94 - 1816235 AACUUGA-UGAGAUUUCUGUUGUUUUUAGAAGCCUGAUGUUAUUUA-AGCG--UCCAGUGAUAUCAAUCCGAUGU--UGUUUGCAUUGUUUUGUAAUUAU-------------------- .......-..((..(((((.......)))))..))((((((.....-))))--))..(((((..(((..((((((--.....))))))..)))..)))))-------------------- ( -17.20, z-score = -1.24, R) >consensus AACUUGAUUGCGA__UGUGUGGUGUCUAUAAAUUGUGAAUUACUCAUGGCA__UGCAUUUGCAUGUCUCUGCCUUAAGGUUUUCCUCCGAAGGCGAUCCGCAUUGCAAAUGCAUUACGCU ................(((((((.........(..(((.....)))..).....((((((((((((...........((.((.((......)).)).))))).)))))))))))))))). (-12.64 = -14.73 + 2.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:39 2011