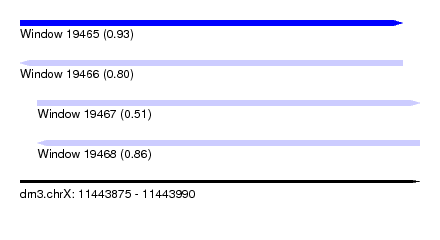
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,443,875 – 11,443,990 |
| Length | 115 |
| Max. P | 0.931959 |

| Location | 11,443,875 – 11,443,985 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 62.43 |
| Shannon entropy | 0.62447 |
| G+C content | 0.53928 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.931959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11443875 110 + 22422827 UGCCCAAAGGGGGGCAGGAGAACCCUUCCCCCCCUGAAUGACAAGAGAAGCAGCUGGCCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCUUCUUUCCUCUGGCCACC--------- .(((....((((((.(((.....))).))))))......((..((((((((.((((((..(((....)))..))))))((((....))))))))))))..)).)))....--------- ( -45.00, z-score = -2.94, R) >droWil1.scaffold_180777 125556 90 + 4753960 ---------AGUUGGAGGAGGAGGA--------AAAGGAGGCAAGAGUAGCAAAUGGUUAACUAUUUGGUAUGCCAUCUUUGUCACCAAUAUCAAUAUUAUUCUCUC------------ ---------....((((((((.(..--------((((..((((....((.(((((((....))))))).))))))..))))..).))((((....)))).)))))).------------ ( -23.70, z-score = -1.93, R) >droAna3.scaffold_13047 190692 104 + 1816235 ----UCCGGAGUGGUCGGGAGACGC----------GAGCAGU-GCAGGAGCAGCUGGGCAACUAUUUGGUAUGCCAGCGCUGUCACCAAUGCUUCUUUUCCCAGCCGCUUCUGGCCACG ----.((((((((((.((((((.(.----------(((((.(-(..(..(((((..(((((((....))).))))...)))))..))).)))))).)))))).))))).)))))..... ( -44.90, z-score = -1.88, R) >droEre2.scaffold_4690 15030383 101 - 18748788 -UCCCACAGGG--GCAGGAGGAGCC------CACUGAACGGUCGGAGAAGCAGCUGGUCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCUUCUUUUCCCCGGCCACC--------- -.....(((((--((.......)))------).)))...((((((((((((.((((((..(((....)))..))))))((((....)))))))))).....))))))...--------- ( -40.20, z-score = -2.24, R) >droYak2.chrX 11052191 96 - 21770863 -UCCCAAAGGG--GCAGGAGGAGGC------CCCUGAACGA-----GAAGCAGCUGGUCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCCUCUUUCCUCCGGCCACC--------- -.......(((--((.(((((((((------...((...((-----.((..(((((((..(((....)))..))))))))).))..))..)))....)))))).))).))--------- ( -33.70, z-score = -0.73, R) >consensus __CCCAAAGGG__GCAGGAGGAGGC______C_CUGAACGGC_AGAGAAGCAGCUGGUCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCUUCUUUUCUCCGGCCACC_________ .............((.((((((.......................((((((.((((((..(((....)))..))))))((((....)))))))))).)))))).))............. (-14.15 = -14.83 + 0.68)

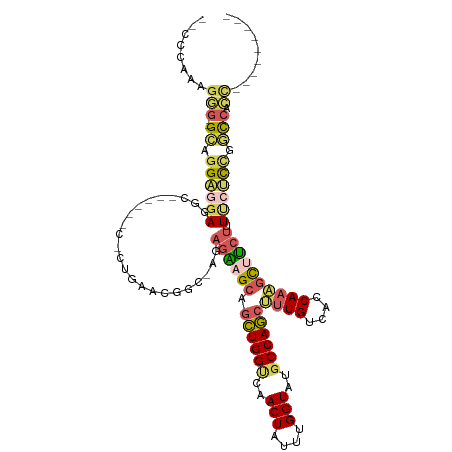
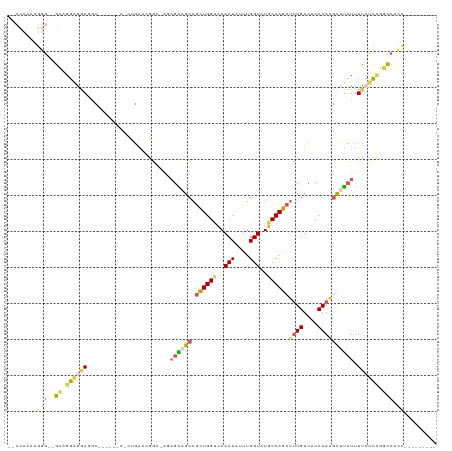
| Location | 11,443,875 – 11,443,985 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 62.43 |
| Shannon entropy | 0.62447 |
| G+C content | 0.53928 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -16.00 |
| Energy contribution | -15.68 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11443875 110 - 22422827 ---------GGUGGCCAGAGGAAAGAAGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGGCCAGCUGCUUCUCUUGUCAUUCAGGGGGGGAAGGGUUCUCCUGCCCCCCUUUGGGCA ---------....(((.((.((.((((((.(((....)))(((((((..((......))..))))))))))))).)).))...(((((((((..(((....)))..)))))))))))). ( -53.70, z-score = -3.01, R) >droWil1.scaffold_180777 125556 90 - 4753960 ------------GAGAGAAUAAUAUUGAUAUUGGUGACAAAGAUGGCAUACCAAAUAGUUAACCAUUUGCUACUCUUGCCUCCUUU--------UCCUCCUCCUCCAACU--------- ------------(((.(((...........(((....))).((.((((((.(((((.(....).))))).))....))))))...)--------)))))...........--------- ( -13.50, z-score = -0.48, R) >droAna3.scaffold_13047 190692 104 - 1816235 CGUGGCCAGAAGCGGCUGGGAAAAGAAGCAUUGGUGACAGCGCUGGCAUACCAAAUAGUUGCCCAGCUGCUCCUGC-ACUGCUC----------GCGUCUCCCGACCACUCCGGA---- (((((((((.((((((((((......(((.(((....))).)))((....)).........)))))))))).))).-...)).)----------)))....(((.......))).---- ( -39.80, z-score = -1.06, R) >droEre2.scaffold_4690 15030383 101 + 18748788 ---------GGUGGCCGGGGAAAAGAAGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGACCAGCUGCUUCUCCGACCGUUCAGUG------GGCUCCUCCUGC--CCCUGUGGGA- ---------((.(((.(((((..((((((.(((....)))((((((((.((......)))).))))))))))))((.((......)).------)).))).)).))--))).......- ( -38.00, z-score = -0.79, R) >droYak2.chrX 11052191 96 + 21770863 ---------GGUGGCCGGAGGAAAGAGGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGACCAGCUGCUUC-----UCGUUCAGGG------GCCUCCUCCUGC--CCCUUUGGGA- ---------((.(((.((((((.((((((.(((....)))((((((((.((......)))).)))))))))))-----).(((....)------)).)))))).))--))).......- ( -40.30, z-score = -1.40, R) >consensus _________GGUGGCCGGAGAAAAGAAGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGACCAGCUGCUUCUCC_ACCGUUCAG_G______GCCUCCUCCUGC__CCCUGUGGG__ .................((....((((((.(((....)))((((((...((......))...))))))))))))....))....................................... (-16.00 = -15.68 + -0.32)

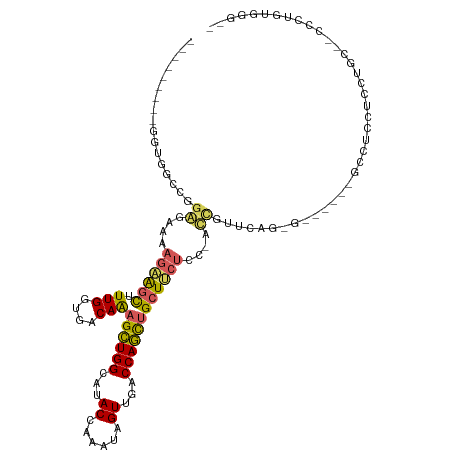
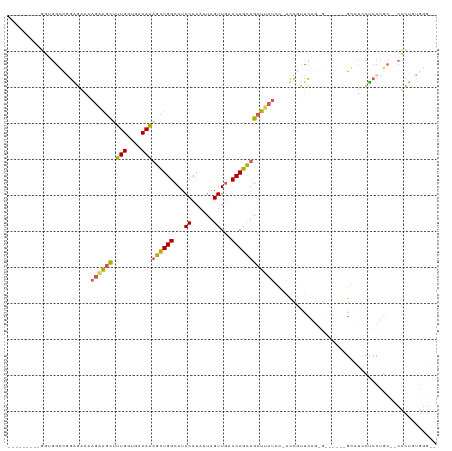
| Location | 11,443,880 – 11,443,990 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.08 |
| Shannon entropy | 0.54936 |
| G+C content | 0.53540 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11443880 110 + 22422827 AAAGGGGGGCAGGAGAACCCUUCCCCCCCUGAAUGACAAGAGAAGCAGCUGGCCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCUUCUUUCC---------UCUGGCCACCGCUCU ...((((((.(((.....))).))))))......((..((((((((.((((((..(((....)))..))))))((((....))))))))))))..---------)).(((....))).. ( -41.90, z-score = -2.01, R) >droWil1.scaffold_180777 125561 85 + 4753960 ---GAGGAGGAGGAAAAGGA-----------GGCAA---GAGUAGCAAAUGGUUAACUAUUUGGUAUGCCAUCUUUGUCACCAAUAUCAAUAUUA--------UUCUCUC--------- ---(((((((.(..((((..-----------((((.---...((.(((((((....))))))).))))))..))))..).))((((....)))).--------)))))..--------- ( -23.00, z-score = -2.31, R) >droAna3.scaffold_13047 190697 104 + 1816235 ----AGUGGUCGGGAGACGC----------GAGCAGU-GCAGGAGCAGCUGGGCAACUAUUUGGUAUGCCAGCGCUGUCACCAAUGCUUCUUUUCCCAGCCGCUUCUGGCCACGGCUCU ----.(((.((....)))))----------((((.((-((((((((.((((((........(((....)))(((.((....)).))).......)))))).)))))))..))).)))). ( -41.90, z-score = -0.74, R) >droEre2.scaffold_4690 15030388 101 - 18748788 ---CAGGGGCAGGAGGAGCC------CACUGAACGGUCGGAGAAGCAGCUGGUCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCUUCUUUUC---------CCCGGCCACCGCUCU ---(((((((.......)))------).)))...((((((((((((.((((((..(((....)))..))))))((((....))))))))))....---------.))))))........ ( -39.70, z-score = -1.74, R) >droYak2.chrX 11052196 96 - 21770863 ---AAGGGGCAGGAGGAGGC------CCCUGAACGA-----GAAGCAGCUGGUCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCCUCUUUCC---------UCCGGCCACCGCUCU ---..(((((.(((((((((------...((...((-----.((..(((((((..(((....)))..))))))))).))..))..)))....)))---------))).))).))..... ( -35.40, z-score = -0.80, R) >consensus ___GAGGGGCAGGAGGAGGC______C_CUGAACGA___GAGAAGCAGCUGGUCAACUAUUUGGUAUGCCAGCUUUGUCACCAAAGCUUCUUUUC_________UCUGGCCACCGCUCU .......(((.(((.................................((((((..(((....)))..))))))((((....))))...................))).)))........ (-14.38 = -14.10 + -0.28)

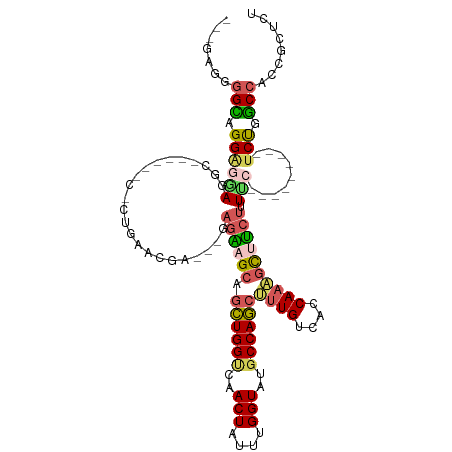
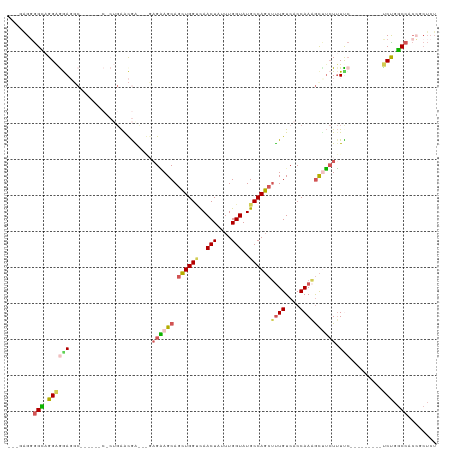
| Location | 11,443,880 – 11,443,990 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.08 |
| Shannon entropy | 0.54936 |
| G+C content | 0.53540 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -15.64 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11443880 110 - 22422827 AGAGCGGUGGCCAGA---------GGAAAGAAGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGGCCAGCUGCUUCUCUUGUCAUUCAGGGGGGGAAGGGUUCUCCUGCCCCCCUUU .(((((((((((((.---------.......(((((((....)))))))((....))......))))))).))))))............((((((((.(((.....))).)))))))). ( -50.60, z-score = -2.64, R) >droWil1.scaffold_180777 125561 85 - 4753960 ---------GAGAGAA--------UAAUAUUGAUAUUGGUGACAAAGAUGGCAUACCAAAUAGUUAACCAUUUGCUACUC---UUGCC-----------UCCUUUUCCUCCUCCUC--- ---------(((.(((--------...........(((....))).((.((((((.(((((.(....).))))).))...---.))))-----------))...))))))......--- ( -13.50, z-score = -0.44, R) >droAna3.scaffold_13047 190697 104 - 1816235 AGAGCCGUGGCCAGAAGCGGCUGGGAAAAGAAGCAUUGGUGACAGCGCUGGCAUACCAAAUAGUUGCCCAGCUGCUCCUGC-ACUGCUC----------GCGUCUCCCGACCACU---- .((((.(((..(((.((((((((((......(((.(((....))).)))((....)).........)))))))))).))))-)).))))----------(.(((....))))...---- ( -43.70, z-score = -2.24, R) >droEre2.scaffold_4690 15030388 101 + 18748788 AGAGCGGUGGCCGGG---------GAAAAGAAGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGACCAGCUGCUUCUCCGACCGUUCAGUG------GGCUCCUCCUGCCCCUG--- .(((((((...((((---------(((.((.(((((((....)))))))((((.((......)))).))..))..))))))))))))))((.(------(((.......)))))).--- ( -42.50, z-score = -2.55, R) >droYak2.chrX 11052196 96 + 21770863 AGAGCGGUGGCCGGA---------GGAAAGAGGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGACCAGCUGCUUC-----UCGUUCAGGG------GCCUCCUCCUGCCCCUU--- .....((.(((.(((---------(((.((((((.(((....)))((((((((.((......)))).)))))))))))-----).(((....)------)).)))))).)))))..--- ( -40.60, z-score = -1.76, R) >consensus AGAGCGGUGGCCAGA_________GAAAAGAAGCUUUGGUGACAAAGCUGGCAUACCAAAUAGUUGACCAGCUGCUUCUC___UCGUUCAG_G______GCCUCCUCCUGCCCCUC___ .(((((((((.(((.................(((((((....)))))))((....))......))).))).)))))).......................................... (-15.64 = -17.32 + 1.68)

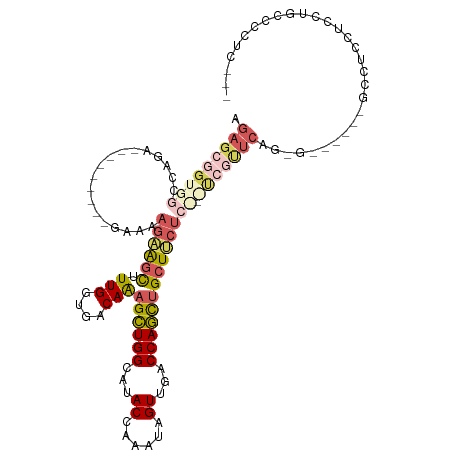
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:37 2011