| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,443,614 – 11,443,778 |
| Length | 164 |
| Max. P | 0.553749 |

| Location | 11,443,614 – 11,443,778 |
|---|---|
| Length | 164 |
| Sequences | 5 |
| Columns | 183 |
| Reading direction | reverse |
| Mean pairwise identity | 62.70 |
| Shannon entropy | 0.64020 |
| G+C content | 0.32187 |
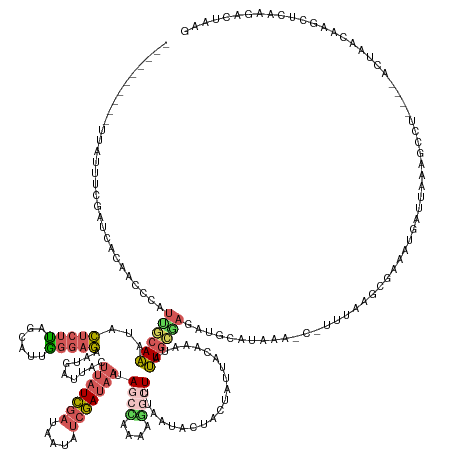
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -8.84 |
| Energy contribution | -10.12 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
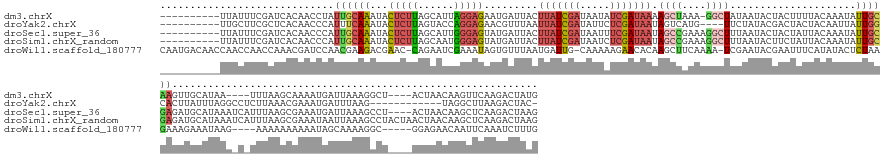
>dm3.chrX 11443614 164 - 22422827 ----------UUAUUUCGAUCACAACCUAUUGCAAAUACUCUUAGCAUUAGGAGAAUGAUUACUUAUCGAUAAUAUCGAUAAAAGCUAAA-GGCUAUAAUACUACUUUUACAAAUAUUGCAAGUUGCAUAA----UUUAAGCAAAAUGAUUAAAGGCU----ACUAACAAGUUCAAGACUAUG ----------...(((..((((.......((((((...((((((....))))))..((.....((((((((...)))))))).(((....-.)))...............))....)))))).((((....----.....))))..))))..)))...----..................... ( -23.90, z-score = 0.01, R) >droYak2.chrX 11051903 156 + 21770863 ----------UUGCUUCGCUCACAACCCAUUUCAAAUACUCUUAGUACCAGGAGAACGUUUAAUUAUCGAUAUUCUCGAUAAUAGUCAUG----UUCUAUACGACUACUACAAUUAUUGGCACUUAUUUAGGCCUCUUAAACGAAAUGAUUUAAG------------UAGGCUUAAGACUAC- ----------.(((............................(((((...(.(((((((...((((((((.....))))))))....)))----))))...)...)))))((.....)))))....(((((((((((((((........))))))------------.))))))))).....- ( -31.90, z-score = -2.21, R) >droSec1.super_36 157801 169 - 449511 ----------UUAUUUCGAUCACAACCCAUUGCAAAUACUCUUAGCAUUGGGAGUAUGAUUACUUAUCGAUAAUUUCGAUAAUAGCCGAAAGGCUUUAAUACUACUAUUACAAAUAUUGCGAGAUGCAUAAAUCAUUUAAGCGAAAUGAUUAAAGCCU----ACUAACAAGCUCAAGACUAAG ----------..((((((.......((((.(((...........))).))))(((((......(((((((.....))))))).((((....))))...)))))................))))))((...((((((((.....))))))))...))..----..................... ( -37.90, z-score = -3.51, R) >droSim1.chrX_random 3205758 173 - 5698898 ----------UUAUUUCGAUCACAACCCAUUGCAAAUACUCUUAGCAAUGGGAGUAUGAUUACUUAUCGAUAAUCUCGAUAAUAGCCGAAAGGCUUUAAUACUUCUAUUACAAAUAUUGCGAGAUGCAUAAAUCAUUUAAGCGAAAUAAUUAAAGCCUACUAACUAACAAGCUCAAGACUAAG ----------((((((((.((.(((((((((((...........))))))))(((((......(((((((.....))))))).((((....))))...))))).............))).))(((......))).......)))))))).................................. ( -39.10, z-score = -3.95, R) >droWil1.scaffold_180777 125264 171 - 4753960 CAAUGACAACCAACCAACCAAACGAUCCAACGAAGACGAAC-CAGAAUCGAAAUAGUGUUUAAUGAUUG-CAAAAAGAACACAAGCUUCAAAA-UCGAAUACGAAUUUCAUAUACUCUAAGAAAGAAAUAAG----AAAAAAAAAAUAGCAAAAGGC-----GGAGAACAAUUCAAAUCUUUG ..((((.........................((((.(((..-.....))).....((((((........-......))))))...))))....-(((....)))...))))..........(((((......----............((.....))-----(((......)))...))))). ( -16.24, z-score = 0.92, R) >consensus __________UUAUUUCGAUCACAACCCAUUGCAAAUACUCUUAGCAUUGGGAGAAUGAUUACUUAUCGAUAAUAUCGAUAAUAGCCAAAAGGCUUUAAUACUACUAUUACAAAUAUUGCGAGAUGCAUAAA_C_UUUAAGCGAAAUGAUUAAAGCCU____ACUAACAAGCUCAAGACUAAG .............................((((((...(((((......))))).........(((((((.....))))))).((((....)))).....................))))))............................................................. ( -8.84 = -10.12 + 1.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:34 2011