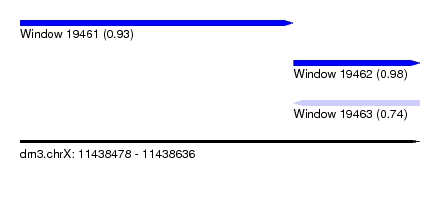
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,438,478 – 11,438,636 |
| Length | 158 |
| Max. P | 0.982812 |

| Location | 11,438,478 – 11,438,586 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Shannon entropy | 0.29326 |
| G+C content | 0.47869 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -26.77 |
| Energy contribution | -25.13 |
| Covariance contribution | -1.65 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
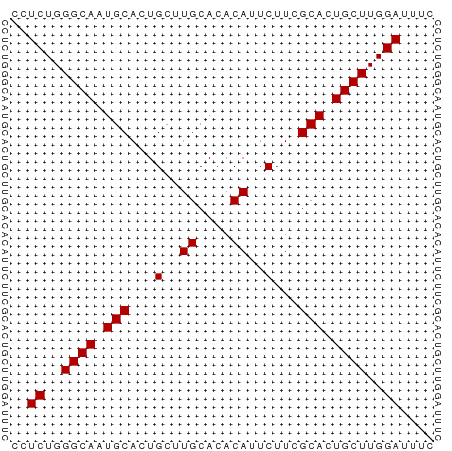
>dm3.chrX 11438478 108 + 22422827 CUUCUCCUUCAUAUA-UGUAUGUCGUA------GUGUAUGCAUGUGUGAGUACGAUAGUCUGUCUCUUUUGCACGACCGCAGGCUGUCUCUUUUGCGCGGCAUUCUUGCAUUUCU .......((((((((-((((((.....------...))))))))))))))...((((((((((.((........))..))))))))))...........(((....)))...... ( -29.70, z-score = -1.77, R) >droYak2.chrX 11046857 115 - 21770863 CUGCUCCUCCAUACAACAUAUACCAUAUAUGUAGCAUGUGCAUGUGUAAGUACGACGGUCUGUCUCUUUCGCUCGCCCGCAGGCUGUCUCUUUUGCGCGGCAUUCUUGCAUUUCU ...............(((((((...))))))).(((.((((.((((((((...((((((((((...............))))))))))...))))))))))))...)))...... ( -33.56, z-score = -2.63, R) >droEre2.scaffold_4690 15025183 109 - 18748788 CUGCUCUGCUCCUC--CAUGUACCAUAUAC----CAUAUGCAUGUGUAAGUACGGCGGUCUGUCUCUUUUGCACGCCCGCAGGCUGUCUCUUUUAUGCGGCAUUCUUGCAUUUCU ..(((.((((...(--((((((........----....)))))).)..)))).)))(((.(((.......))).))).(((((.((((.(......).))))..)))))...... ( -24.30, z-score = 0.02, R) >consensus CUGCUCCUCCAUACA_CAUAUACCAUAUA____GCAUAUGCAUGUGUAAGUACGACGGUCUGUCUCUUUUGCACGCCCGCAGGCUGUCUCUUUUGCGCGGCAUUCUUGCAUUUCU .................................(((.((((.((((((((...((((((((((...............))))))))))...))))))))))))...)))...... (-26.77 = -25.13 + -1.65)

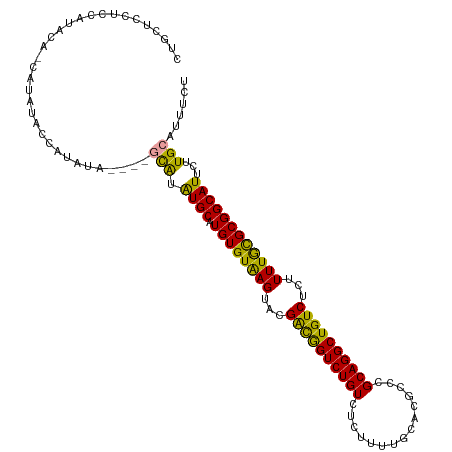
| Location | 11,438,586 – 11,438,636 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 50 |
| Reading direction | forward |
| Mean pairwise identity | 99.20 |
| Shannon entropy | 0.01444 |
| G+C content | 0.51600 |
| Mean single sequence MFE | -13.00 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
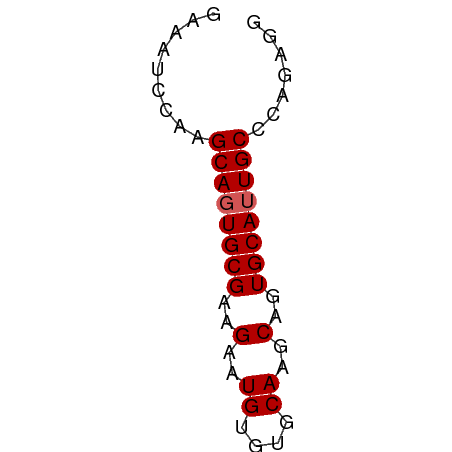
>dm3.chrX 11438586 50 + 22422827 CCUCUGGGCAAUGCACUGCUUGCACACAUUCUUCGCACUGCUUGGAUUUC ..((..((((.(((...(..((....))..)...))).))))..)).... ( -13.00, z-score = -2.30, R) >droSim1.chrX 8816691 50 + 17042790 CCUCUGGGCAAUGCACUGCUUGCACACAUUCUUCGCACUGCUUGGAUUUC ..((..((((.(((...(..((....))..)...))).))))..)).... ( -13.00, z-score = -2.30, R) >droSec1.super_36 152975 50 + 449511 CCUCUGGGCAAUGCACUGCUUGCACACAUUCUUCGCACUGCUUGGAUUUC ..((..((((.(((...(..((....))..)...))).))))..)).... ( -13.00, z-score = -2.30, R) >droYak2.chrX 11046972 50 - 21770863 CCUCUGGGCAAUGCACUGCUUGCACACAUUCUUCGCACUGCUUGGAUUUC ..((..((((.(((...(..((....))..)...))).))))..)).... ( -13.00, z-score = -2.30, R) >droEre2.scaffold_4690 15025292 50 - 18748788 CCUCUGGGCAAUGCACUGCUUGCACACAUUCUUCGCAAUGCUUGGAUUUC ..((..((((.(((...(..((....))..)...))).))))..)).... ( -13.00, z-score = -1.99, R) >consensus CCUCUGGGCAAUGCACUGCUUGCACACAUUCUUCGCACUGCUUGGAUUUC ..((..((((.(((...(..((....))..)...))).))))..)).... (-13.00 = -13.00 + 0.00)

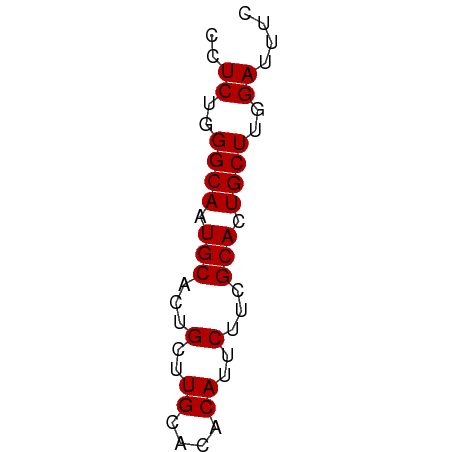
| Location | 11,438,586 – 11,438,636 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 50 |
| Reading direction | reverse |
| Mean pairwise identity | 99.20 |
| Shannon entropy | 0.01444 |
| G+C content | 0.51600 |
| Mean single sequence MFE | -12.13 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
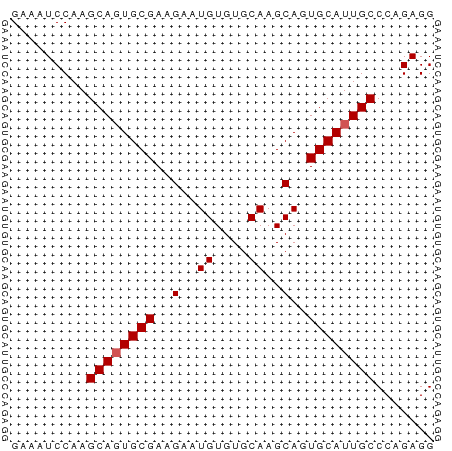
>dm3.chrX 11438586 50 - 22422827 GAAAUCCAAGCAGUGCGAAGAAUGUGUGCAAGCAGUGCAUUGCCCAGAGG .........((((((((..(..((....))..)..))))))))....... ( -12.60, z-score = -1.46, R) >droSim1.chrX 8816691 50 - 17042790 GAAAUCCAAGCAGUGCGAAGAAUGUGUGCAAGCAGUGCAUUGCCCAGAGG .........((((((((..(..((....))..)..))))))))....... ( -12.60, z-score = -1.46, R) >droSec1.super_36 152975 50 - 449511 GAAAUCCAAGCAGUGCGAAGAAUGUGUGCAAGCAGUGCAUUGCCCAGAGG .........((((((((..(..((....))..)..))))))))....... ( -12.60, z-score = -1.46, R) >droYak2.chrX 11046972 50 + 21770863 GAAAUCCAAGCAGUGCGAAGAAUGUGUGCAAGCAGUGCAUUGCCCAGAGG .........((((((((..(..((....))..)..))))))))....... ( -12.60, z-score = -1.46, R) >droEre2.scaffold_4690 15025292 50 + 18748788 GAAAUCCAAGCAUUGCGAAGAAUGUGUGCAAGCAGUGCAUUGCCCAGAGG ......((((((((((...............))))))).)))........ ( -10.26, z-score = -0.20, R) >consensus GAAAUCCAAGCAGUGCGAAGAAUGUGUGCAAGCAGUGCAUUGCCCAGAGG .........((((((((..(..((....))..)..))))))))....... (-12.00 = -12.20 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:33 2011