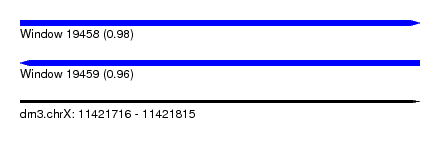
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,421,716 – 11,421,815 |
| Length | 99 |
| Max. P | 0.981672 |

| Location | 11,421,716 – 11,421,815 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.05 |
| Shannon entropy | 0.47514 |
| G+C content | 0.40781 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -10.55 |
| Energy contribution | -11.58 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
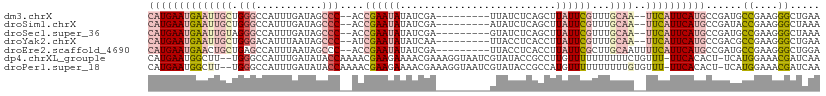
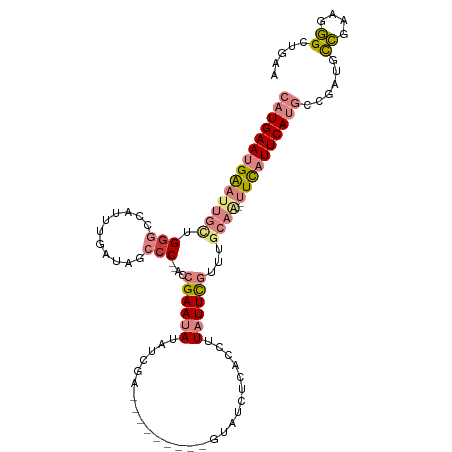
>dm3.chrX 11421716 99 + 22422827 UUCAGCCCUUCGGCAUCGGCAUGAAUGAA--UUGCAAACGAAUAAGCUGAGAUAA---------UCGAUAUAUUCGGU--GGGCUAUCAAAUGGCCCAGCAAUUCAUUCAUG ....(((....))).....((((((((((--((((...((((((.(.(((.....---------))).).)))))).(--(((((((...)))))))))))))))))))))) ( -40.40, z-score = -5.39, R) >droSim1.chrX 8806314 99 + 17042790 UUUAGCCCUUCGGUAUCGGCAUGAAUGAA--UUGCAAACGAAUAAGCUGAGAUAU---------UCGAUAUAUUCGGU--GGGCUAUCAAAUGGCCCAGCAAUUCAUUCAUG ....((((....)....)))(((((((((--((((...((((((.(.((((...)---------))).).)))))).(--(((((((...))))))))))))))))))))). ( -37.80, z-score = -4.92, R) >droSec1.super_36 143283 99 + 449511 UUUAGCCCUUCGGCAUCGGCAUGAAUGAA--UUGCAAACGAAUAAGCUGAGAUAC---------UCGAUAUAUUCGGU--GGGCUAUCAAAUGGCCCUACAAUUCAUUCAUG ....(((....))).....((((((((((--(((....((((((.(.((((...)---------))).).))))))..--(((((((...)))))))..))))))))))))) ( -35.50, z-score = -4.63, R) >droYak2.chrX 11037179 99 - 21770863 UUCAGCCCUUCGGCGUCGGCAUGAAUGAA--UUGCAAACGAAUAAGGUGAGGUAA---------UUGAUAUAUUCGAU--GGGCUAUUAAAUGUCCCAGCAAUUCAUUCAUG ....(((....))).....((((((((((--((((...((((((..((.((....---------)).)).)))))).(--((((........).)))))))))))))))))) ( -33.60, z-score = -3.62, R) >droEre2.scaffold_4690 15013330 101 - 18748788 UCCAGCCCUUCGGCAUCGGCAUGAAUGAAAAUUGCAAGCGAAUAAGGUGAGGUAA---------UCGAUAUAUUCGGU--GGGCUAUUAAAUGGCUCAGCAGUUCAUUCAUG ....(((....))).....(((((((((..(((((...((((((...(((.....---------)))...)))))).(--(((((((...)))))))))))))))))))))) ( -33.50, z-score = -2.89, R) >dp4.chrXL_group1e 1752604 108 + 12523060 UUGAUCGUUUCCAUGA-AGUGUGAA-AAACAGAAAAAAAAAACAAGGCGGUAUACGAUUACCUUUCGUUUUCUUCGUUUUGGUAUAUCAAAUGGCCCA--AAGCCAUUCAUG ...........(((((-(.(((...-..)))...........((((((((...((((.......)))).....))))))))..........((((...--..)))))))))) ( -20.90, z-score = -0.72, R) >droPer1.super_18 591884 108 + 1952607 UUGAUCGUUUCCAUGA-AGUGUGAA-AAACACAAAAAAAAAACAUGGCGGUAUACGAUUACCUUUCGUUUUCUUCGUUUUGGUAUAUCAAAUGGCCCA--AAGCCAUUCAUG ...........(((((-(((((...-..))))............((((((((......))))...............(((((..(((...)))..)))--)))))))))))) ( -22.50, z-score = -1.21, R) >consensus UUCAGCCCUUCGGCAUCGGCAUGAAUGAA__UUGCAAACGAAUAAGGUGAGAUAA_________UCGAUAUAUUCGGU__GGGCUAUCAAAUGGCCCAGCAAUUCAUUCAUG ....(((....))).....(((((((((..........((((((...(((..............)))...))))))....(((...........)))......))))))))) (-10.55 = -11.58 + 1.03)

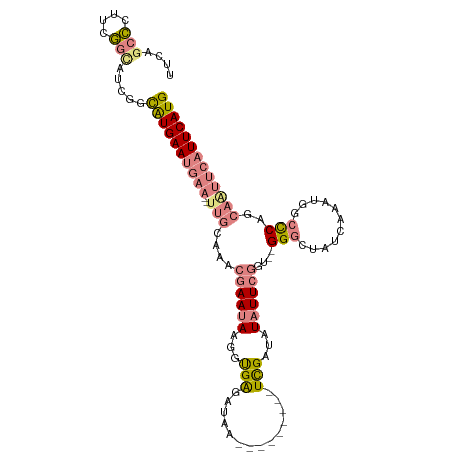
| Location | 11,421,716 – 11,421,815 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.05 |
| Shannon entropy | 0.47514 |
| G+C content | 0.40781 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -9.88 |
| Energy contribution | -11.78 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11421716 99 - 22422827 CAUGAAUGAAUUGCUGGGCCAUUUGAUAGCCC--ACCGAAUAUAUCGA---------UUAUCUCAGCUUAUUCGUUUGCAA--UUCAUUCAUGCCGAUGCCGAAGGGCUGAA (((((((((((((((((((.........))))--).((((((....((---------.....))....))))))...))))--)))))))))).....(((....))).... ( -36.60, z-score = -4.06, R) >droSim1.chrX 8806314 99 - 17042790 CAUGAAUGAAUUGCUGGGCCAUUUGAUAGCCC--ACCGAAUAUAUCGA---------AUAUCUCAGCUUAUUCGUUUGCAA--UUCAUUCAUGCCGAUACCGAAGGGCUAAA (((((((((((((((((((.........))))--)..........(((---------(((........))))))...))))--))))))))))......((....))..... ( -33.40, z-score = -4.07, R) >droSec1.super_36 143283 99 - 449511 CAUGAAUGAAUUGUAGGGCCAUUUGAUAGCCC--ACCGAAUAUAUCGA---------GUAUCUCAGCUUAUUCGUUUGCAA--UUCAUUCAUGCCGAUGCCGAAGGGCUAAA (((((((((((((((((((.........))))--..((((((....((---------(...)))....))))))..)))))--)))))))))).....(((....))).... ( -37.40, z-score = -4.88, R) >droYak2.chrX 11037179 99 + 21770863 CAUGAAUGAAUUGCUGGGACAUUUAAUAGCCC--AUCGAAUAUAUCAA---------UUACCUCACCUUAUUCGUUUGCAA--UUCAUUCAUGCCGACGCCGAAGGGCUGAA ((((((((((((((((((.(........))))--).((((((......---------...........))))))...))))--)))))))))).....(((....))).... ( -33.53, z-score = -4.74, R) >droEre2.scaffold_4690 15013330 101 + 18748788 CAUGAAUGAACUGCUGAGCCAUUUAAUAGCCC--ACCGAAUAUAUCGA---------UUACCUCACCUUAUUCGCUUGCAAUUUUCAUUCAUGCCGAUGCCGAAGGGCUGGA ((((((((((.(((.((((.........(...--..)(((((....((---------.....))....))))))))))))...))))))))))(((..(((....)))))). ( -25.20, z-score = -1.91, R) >dp4.chrXL_group1e 1752604 108 - 12523060 CAUGAAUGGCUU--UGGGCCAUUUGAUAUACCAAAACGAAGAAAACGAAAGGUAAUCGUAUACCGCCUUGUUUUUUUUUUCUGUUU-UUCACACU-UCAUGGAAACGAUCAA ...(((((((..--...)))))))(((.....((((((((((((((((..((((......))))...)))))))))))....))))-).......-...((....))))).. ( -23.90, z-score = -1.46, R) >droPer1.super_18 591884 108 - 1952607 CAUGAAUGGCUU--UGGGCCAUUUGAUAUACCAAAACGAAGAAAACGAAAGGUAAUCGUAUACCGCCAUGUUUUUUUUUUGUGUUU-UUCACACU-UCAUGGAAACGAUCAA ...(((((((..--...)))))))(((...(((....((((((((((...((((......))))....)))))))))).((((...-..))))..-...))).....))).. ( -24.60, z-score = -1.28, R) >consensus CAUGAAUGAAUUGCUGGGCCAUUUGAUAGCCC__ACCGAAUAUAUCGA_________GUAUCUCACCUUAUUCGUUUGCAA__UUCAUUCAUGCCGAUGCCGAAGGGCUGAA ((((((((((((((.(((...........)))....((((((..........................))))))...))))..))))))))))......((....))..... ( -9.88 = -11.78 + 1.90)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:30 2011