| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,406,484 – 11,406,575 |
| Length | 91 |
| Max. P | 0.837482 |

| Location | 11,406,484 – 11,406,575 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.04 |
| Shannon entropy | 0.43350 |
| G+C content | 0.50172 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -11.65 |
| Energy contribution | -12.24 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837482 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 11406484 91 + 22422827 CAACACAUAUGCACACAUAUGCAUACACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAAC .......((((((......))))))...........((((((((((((..................))))...)))).))))......... ( -18.37, z-score = -1.73, R) >droSim1.chrX_random 3187386 89 + 5698898 --CAACACAUGCACACAUGUGCAUAUACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAAC --.......(((...((((((......))))))...))).((((((((.........(((....))).....((....))))))))))... ( -21.90, z-score = -1.97, R) >droSec1.super_36 128297 89 + 449511 --CAACACUUGCACACAUGUGCAUACACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACACACAAC --......((((...((((((......))))))...))))((((((((.........(((....))).....((....))))))))))... ( -23.00, z-score = -2.42, R) >droYak2.chrX 11021910 77 - 21770863 --------UAACACACACAUGCAUAUGCACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACA------ --------.......((..(((....)))..))...((((((((((((..................))))...)))).))))...------ ( -16.47, z-score = -1.07, R) >droEre2.scaffold_4690 14998150 77 - 18748788 --------UAACACACACAUGCAUAUACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACA------ --------.........((((........))))...((((((((((((..................))))...)))).))))...------ ( -15.77, z-score = -1.65, R) >droVir3.scaffold_12970 2419606 75 + 11907090 ---CGCAUGUGUAAACAAAAACACACACGCGCACGCGCAUUUGUAUGCAACCACCGAAUGACCUUAGCACUCAACACC------------- ---.((((((((........))))....(((....)))......)))).......((.((.......)).))......------------- ( -12.30, z-score = 0.07, R) >consensus ____ACA_UUGCACACAUAUGCAUACACACAUGACCGCGAGUGUGUGCAACCACCAACUGACCUUAGCACUCGACACAUCGCACA______ .........(((((((((.(((..............))).))))))))).......................................... (-11.65 = -12.24 + 0.59)

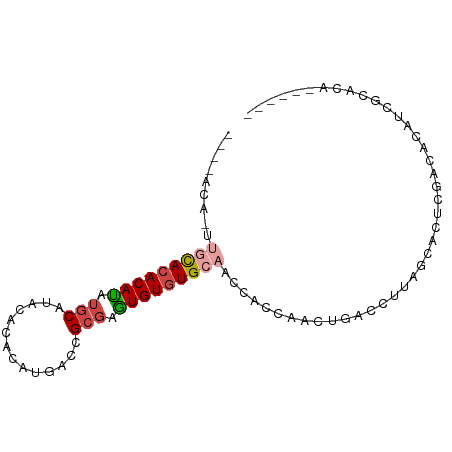
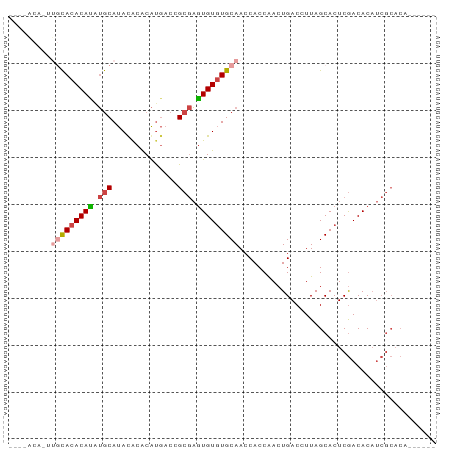
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:27 2011