
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,402,762 – 11,402,819 |
| Length | 57 |
| Max. P | 0.796847 |

| Location | 11,402,762 – 11,402,819 |
|---|---|
| Length | 57 |
| Sequences | 9 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Shannon entropy | 0.35113 |
| G+C content | 0.36827 |
| Mean single sequence MFE | -12.53 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.97 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11402762 57 - 22422827 AGAGAUAUAGAGUC------GUGUAC-UGCAAAAGUUCAAAGUGCUCUAAAGUGCUUUGACUUC ...(((.....)))------......-.....((((.(((((..((....))..))))))))). ( -11.90, z-score = -0.64, R) >droSim1.chrX_random 3181722 57 - 5698898 AGAGAUAUAGAGUC------GUGUAC-UGCAAAAGUUCAAAGUGCUCUAAAGUGCUUUGACUUC ...(((.....)))------......-.....((((.(((((..((....))..))))))))). ( -11.90, z-score = -0.64, R) >droSec1.super_36 124607 57 - 449511 AGAGAUAUAGAGUC------GUGUAC-UGCAAAAGUUCAAAGUGCUCUAAAGUGCUUUGACUUC ...(((.....)))------......-.....((((.(((((..((....))..))))))))). ( -11.90, z-score = -0.64, R) >droYak2.chrX 11017384 57 + 21770863 AGAGAUAUAGAGUC------GUGUAC-UGUAAAAGUUCAAAGUGCUCUAAAGUGCUUUGACUUC .....(((((....------.....)-)))).((((.(((((..((....))..))))))))). ( -12.40, z-score = -1.31, R) >droEre2.scaffold_4690 14994440 57 + 18748788 AGAGAUAUAGAGUC------GUGUAC-UGUAAAAGUUCAAAGUGCUCUAAAGUGCUUUGACUUC .....(((((....------.....)-)))).((((.(((((..((....))..))))))))). ( -12.40, z-score = -1.31, R) >dp4.chrXL_group1e 1727040 57 - 12523060 AGAGACAGAUACUC------GUGUAC-UGUAAAAGUUCAAAGUGCUUUAAAGUGCUUUGACUUC ....((((.(((..------..))))-)))..((((.(((((..((....))..))))))))). ( -16.70, z-score = -2.58, R) >droPer1.super_18 567594 57 - 1952607 AGAGACAGAUACUC------GUGUAC-UGUAAAAGUUCAAAGUGCUUUAAAGUGCUUUGACUUC ....((((.(((..------..))))-)))..((((.(((((..((....))..))))))))). ( -16.70, z-score = -2.58, R) >droWil1.scaffold_180777 87313 64 - 4753960 AGUAAUAAUCAUUCUCUUCUAUAUACACGUAAAAGUUCAAAGUGCUUUAAAGUGCCUUGACUUC ................................((((.(((.(..((....))..).))))))). ( -7.30, z-score = -0.74, R) >droVir3.scaffold_12970 2416287 54 - 11907090 AGAACUGUAGAA--------AUGAA--UGCGGGAAGUCAAAGUACUUUAAAGCACUUUGAACUC ....(((((...--------.....--)))))((..(((((((.((....)).)))))))..)) ( -11.60, z-score = -1.77, R) >consensus AGAGAUAUAGAGUC______GUGUAC_UGUAAAAGUUCAAAGUGCUCUAAAGUGCUUUGACUUC ................................(((.((((((((((....))))))))))))). ( -9.72 = -9.97 + 0.25)

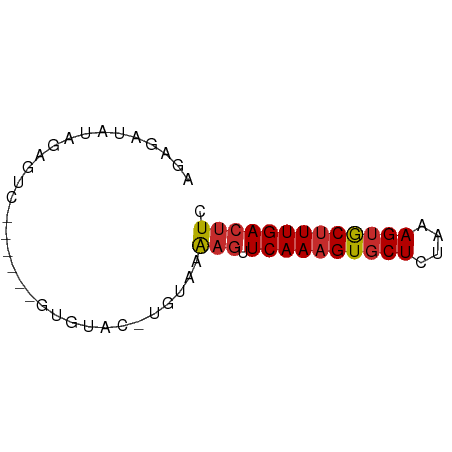
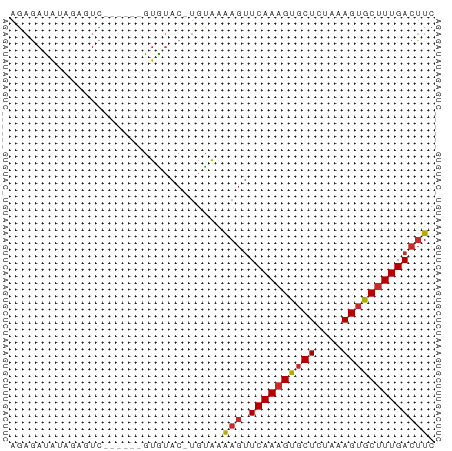
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:26 2011