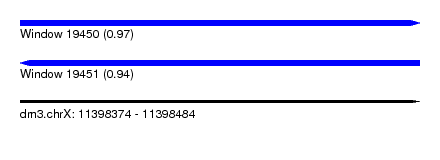
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,398,374 – 11,398,484 |
| Length | 110 |
| Max. P | 0.967902 |

| Location | 11,398,374 – 11,398,484 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Shannon entropy | 0.21949 |
| G+C content | 0.35257 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
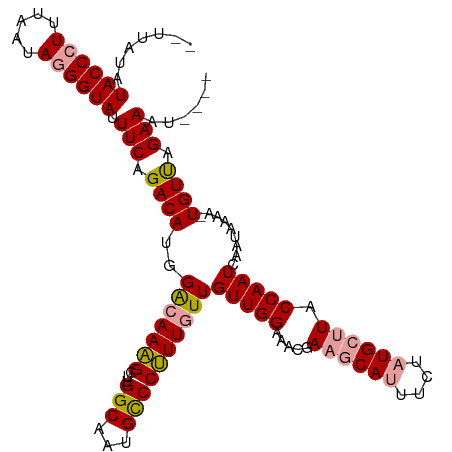
>dm3.chrX 11398374 110 + 22422827 --UUAUAUACCCUUUAAUAGGGUACUUCAGACAUGGACAAAGCUUGGCACUGCCCUUUCUUGUUGGAAACGAAGCAUUUCUAUGCUUACCAAUCAAUAAAAAUGUUAGAAAU---- --.....((((((.....)))))).(((.(((((((.((..((...))..)).))....(((((((.....((((((....)))))).....)))))))..))))).)))..---- ( -25.10, z-score = -1.98, R) >droSec1.super_36 120229 109 + 449511 --UUAUAUACCCUUUAAUAGGGUAUUUCAGACAUGGACAAAGCUUGGCCAUGCCCUUUGUUGUUGGAAACGAAGCAUUUCUAUGCUUACCAAUCAAUAAAU-UGUCCGAAAU---- --....(((((((.....)))))))........(((((((.....(((...))).(((((((((((.....((((((....)))))).)))).))))))))-))))))....---- ( -32.30, z-score = -3.89, R) >droYak2.chrX 11012965 116 - 21770863 UUAUAUAUACCCUUUAAUAAGGUAUUUCAGACAUGGGCAAGGGUUGGCAAUGUCCCUUGUUGUUGGAAACGACACAUUUCUUUGCUUACCAAUUAUUAAAACUGUUAGAAACAAAU ............((((((((((((....(((.(((((((((((..........))))))))((((....)))).))).))).....))))..))))))))................ ( -27.50, z-score = -1.77, R) >consensus __UUAUAUACCCUUUAAUAGGGUAUUUCAGACAUGGACAAAGCUUGGCAAUGCCCUUUGUUGUUGGAAACGAAGCAUUUCUAUGCUUACCAAUCAAUAAAA_UGUUAGAAAU____ .......((((((.....)))))).(((.((((..(((((((...(((...))))))))))(((((.....((((((....)))))).))))).........)))).)))...... (-22.04 = -22.60 + 0.56)

| Location | 11,398,374 – 11,398,484 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Shannon entropy | 0.21949 |
| G+C content | 0.35257 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
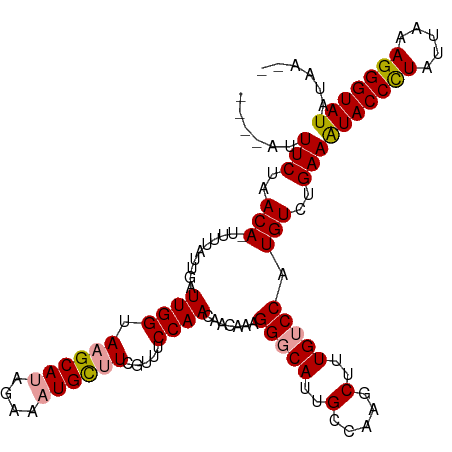
>dm3.chrX 11398374 110 - 22422827 ----AUUUCUAACAUUUUUAUUGAUUGGUAAGCAUAGAAAUGCUUCGUUUCCAACAAGAAAGGGCAGUGCCAAGCUUUGUCCAUGUCUGAAGUACCCUAUUAAAGGGUAUAUAA-- ----..(((..((((.....(((.((((.((((((....)))))).....)))))))....((((((.((...)).))))))))))..)))(((((((.....)))))))....-- ( -30.10, z-score = -2.75, R) >droSec1.super_36 120229 109 - 449511 ----AUUUCGGACA-AUUUAUUGAUUGGUAAGCAUAGAAAUGCUUCGUUUCCAACAACAAAGGGCAUGGCCAAGCUUUGUCCAUGUCUGAAAUACCCUAUUAAAGGGUAUAUAA-- ----..((((((((-.(((.(((.((((.((((((....)))))).....))))))).)))(((((.(((...))).))))).))))))))(((((((.....)))))))....-- ( -37.00, z-score = -4.78, R) >droYak2.chrX 11012965 116 + 21770863 AUUUGUUUCUAACAGUUUUAAUAAUUGGUAAGCAAAGAAAUGUGUCGUUUCCAACAACAAGGGACAUUGCCAACCCUUGCCCAUGUCUGAAAUACCUUAUUAAAGGGUAUAUAUAA .(((((((....(((((.....)))))..)))))))...((((((((((((..(((.((((((..........))))))....)))..))))).((((....))))).)))))).. ( -22.30, z-score = -0.38, R) >consensus ____AUUUCUAACA_UUUUAUUGAUUGGUAAGCAUAGAAAUGCUUCGUUUCCAACAACAAAGGGCAUUGCCAAGCUUUGUCCAUGUCUGAAAUACCCUAUUAAAGGGUAUAUAA__ ......(((..(((..........((((.((((((....)))))).....)))).......(((((..(.....)..))))).)))..)))(((((((.....)))))))...... (-20.73 = -21.07 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:24 2011