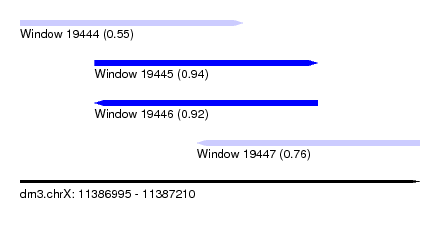
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,386,995 – 11,387,210 |
| Length | 215 |
| Max. P | 0.944600 |

| Location | 11,386,995 – 11,387,115 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.53 |
| Shannon entropy | 0.59116 |
| G+C content | 0.40252 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11386995 120 + 22422827 CUGCCUUUUCUCAGCUUAAAUUAUAAACAAAACUGCACUGUUAUAAACCUAGUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCACAGUGCCUAGCGUUAAACGUCCGGCAACA .((((.............................(((((((.....(((.(((((((............)))))))....))).......)))))))...(((.....)))..))))... ( -23.70, z-score = -0.78, R) >droEre2.scaffold_4690 14978836 119 - 18748788 CUACUUUUUCUCGGUUUGGAUUGAAAAUAAAACUGUACUGUUACCAGCC-AAUGCUGUUAACUGACUUUUGGUAUUUUCUGGUAUAUUCCAAAGUGCCAAUCGUCAAGCGGCUGGCAACA ...........((((((............))))))....(((.((((((-...(((......((((..(((((((((..(((......))))))))))))..))))))))))))).))). ( -33.80, z-score = -3.00, R) >droYak2.chrX 11001463 119 - 21770863 CUAAUUUUUAUCAACUUAGAUAAAUAAUAAACCUGUACUGUUACGAAUC-AGUGCUGUUAACUGGCUUUCGGUAUUUUCCAGUAUAUUCCAAAGUCCCAAGCGUCAUACGGCUGGUCACA ......((((((......)))))).....(((..((((((........)-))))).)))....((((((.((................))))))))(((.((........)))))..... ( -19.89, z-score = 0.02, R) >droSec1.super_36 108831 96 + 449511 -----------------------CUGCCUUUGCUCAGCUAACAU-AACCUAAUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCGCAGUGCCUAGCGUUAAACGCAGGACAACA -----------------------((((.((((....((((....-.(((.((..(((....)))...)).))).......((((((......))))))))))..)))).))))....... ( -20.40, z-score = -0.18, R) >droSim1.chrX 8789040 96 + 17042790 -----------------------CUGCCUUUUCUCAGCUAACAU-AACCUAAUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCGCAGUGCCUAGCGUUAAGCGCGGGACAACA -----------------------(((((((......((((....-.(((.((..(((....)))...)).))).......((((((......))))))))))...))).))))....... ( -20.30, z-score = 0.04, R) >consensus CU___UUUU_UC___UU__AU__AUAACAAAACUGAACUGUUAU_AACCUAAUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCACAGUGCCUAGCGUUAAACGCCGGGCAACA ..............................................(((.(((((((............)))))))....)))..........((((((.(((.....))).)))).)). (-12.24 = -12.08 + -0.16)

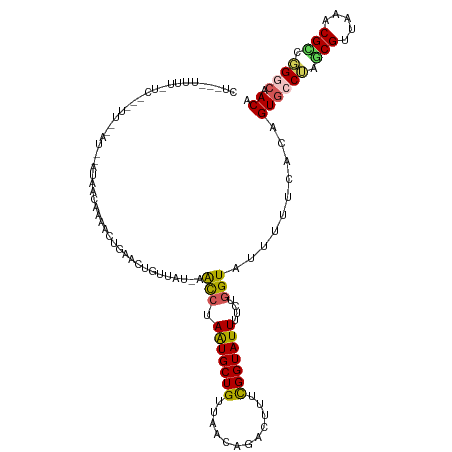
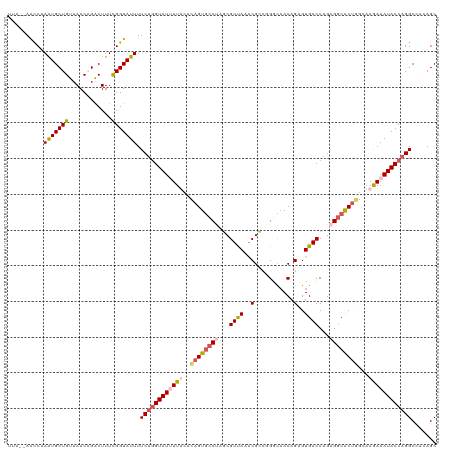
| Location | 11,387,035 – 11,387,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Shannon entropy | 0.32007 |
| G+C content | 0.47289 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -24.84 |
| Energy contribution | -26.04 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11387035 120 + 22422827 UUAUAAACCUAGUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCACAGUGCCUAGCGUUAAACGUCCGGCAACAGAGUGACCAGAGACACUGGCAAGAAAUACCAGGCCCCAGC ......(((.(((.(((....))))))...)))....(((((((((((((((..((((..(((.....)))..)))).....))))((((.....))))...)))))))))))....... ( -34.80, z-score = -2.26, R) >droEre2.scaffold_4690 14978876 119 - 18748788 UUACCAGCC-AAUGCUGUUAACUGACUUUUGGUAUUUUCUGGUAUAUUCCAAAGUGCCAAUCGUCAAGCGGCUGGCAACAGCGUGACCAGAGGCGCCGUCAAGAAAUACCAGGCAAGAGC ......(((-....(((...((((((..(((((((((..(((......))))))))))))..))))....((((....))))))...))).)))(((....(....)....)))...... ( -37.50, z-score = -1.82, R) >droYak2.chrX 11001503 119 - 21770863 UUACGAAUC-AGUGCUGUUAACUGGCUUUCGGUAUUUUCCAGUAUAUUCCAAAGUCCCAAGCGUCAUACGGCUGGUCACAGCGUGACCAGAGGCACUUUCAAAAAAUACCAGGCAAGAGC .........-...(((.((..((((.....((......))..........(((((.((...((.....)).((((((((...)))))))).)).))))).........))))..)).))) ( -29.60, z-score = -0.94, R) >droSec1.super_36 108852 115 + 449511 -----AACCUAAUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCGCAGUGCCUAGCGUUAAACGCAGGACAACAGAGUGACCAGAGGCACUGGCAAGAAAUACCAGGCCUCAGC -----.(((.((..(((....)))...)).)))....((((((((((((.(((((((((.(((.....))).((.((......)).))..))))))).)).))))))))))))....... ( -39.80, z-score = -3.38, R) >droSim1.chrX 8789061 115 + 17042790 -----AACCUAAUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCGCAGUGCCUAGCGUUAAGCGCGGGACAACAGAGUGACCAGAGGCACUGGCAAGAAAUACCAGGCCUCAGC -----.(((.((..(((....)))...)).)))....((((((((((((.(((((((((.(((.....)))((.((......))..))..))))))).)).))))))))))))....... ( -40.70, z-score = -2.96, R) >consensus UUA__AACCUAAUGCUGUUAACAGACUUUCGGUAUUUUCUGGUAUUUUUCACAGUGCCUAGCGUUAAACGCCGGGCAACAGAGUGACCAGAGGCACUGGCAAGAAAUACCAGGCCACAGC ..........(((((((............))))))).((((((((((((..((((((((...((((..(...........)..))))...))))))))...))))))))))))....... (-24.84 = -26.04 + 1.20)

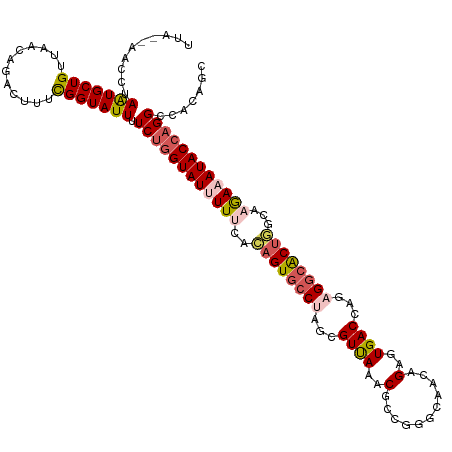
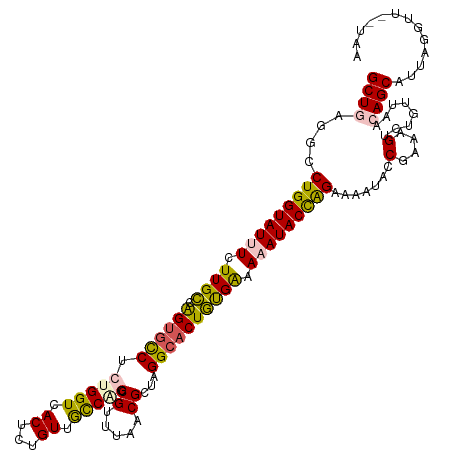
| Location | 11,387,035 – 11,387,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Shannon entropy | 0.32007 |
| G+C content | 0.47289 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11387035 120 - 22422827 GCUGGGGCCUGGUAUUUCUUGCCAGUGUCUCUGGUCACUCUGUUGCCGGACGUUUAACGCUAGGCACUGUGAAAAAUACCAGAAAAUACCGAAAGUCUGUUAACAGCACUAGGUUUAUAA ((((....(((((((((.(..(.((((((((((((.((...)).))))))((.....))...)))))))..).)))))))))..((((.(....)..))))..))))............. ( -36.40, z-score = -1.59, R) >droEre2.scaffold_4690 14978876 119 + 18748788 GCUCUUGCCUGGUAUUUCUUGACGGCGCCUCUGGUCACGCUGUUGCCAGCCGCUUGACGAUUGGCACUUUGGAAUAUACCAGAAAAUACCAAAAGUCAGUUAACAGCAUU-GGCUGGUAA ((....))(((((((((((....((.(((..(.((((.((.((.....)).)).)))).)..))).))..)))).)))))))....(((((...((((((.......)))-)))))))). ( -36.40, z-score = -1.13, R) >droYak2.chrX 11001503 119 + 21770863 GCUCUUGCCUGGUAUUUUUUGAAAGUGCCUCUGGUCACGCUGUGACCAGCCGUAUGACGCUUGGGACUUUGGAAUAUACUGGAAAAUACCGAAAGCCAGUUAACAGCACU-GAUUCGUAA (((.(((.(((((....(((.(((((.((.((((((((...)))))))).((.....))...)).))))).))).....(((......)))...))))).))).)))...-......... ( -33.50, z-score = -1.40, R) >droSec1.super_36 108852 115 - 449511 GCUGAGGCCUGGUAUUUCUUGCCAGUGCCUCUGGUCACUCUGUUGUCCUGCGUUUAACGCUAGGCACUGCGAAAAAUACCAGAAAAUACCGAAAGUCUGUUAACAGCAUUAGGUU----- ((((....(((((((((.((((.(((((((..((.((......)).)).(((.....))).))))))))))).)))))))))..((((.(....)..))))..))))........----- ( -40.80, z-score = -3.57, R) >droSim1.chrX 8789061 115 - 17042790 GCUGAGGCCUGGUAUUUCUUGCCAGUGCCUCUGGUCACUCUGUUGUCCCGCGCUUAACGCUAGGCACUGCGAAAAAUACCAGAAAAUACCGAAAGUCUGUUAACAGCAUUAGGUU----- ((((....(((((((((.((((.(((((((..((.((......)).)).(((.....))).))))))))))).)))))))))..((((.(....)..))))..))))........----- ( -39.90, z-score = -3.08, R) >consensus GCUGAGGCCUGGUAUUUCUUGCCAGUGCCUCUGGUCACUCUGUUGCCAGCCGUUUAACGCUAGGCACUGUGAAAAAUACCAGAAAAUACCGAAAGUCUGUUAACAGCAUUAGGUU__UAA ((((....(((((((((.((((.((((((.(((((.((...)).))))).((.....))...)))))))))).))))))))).......(....)........))))............. (-25.58 = -25.50 + -0.08)

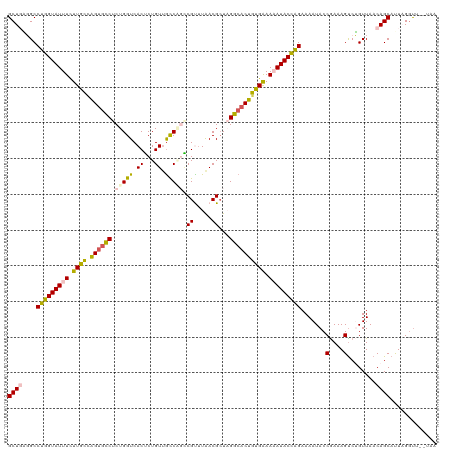
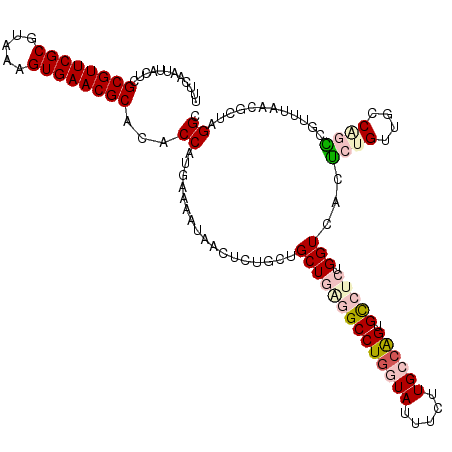
| Location | 11,387,090 – 11,387,210 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Shannon entropy | 0.21067 |
| G+C content | 0.49333 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -25.88 |
| Energy contribution | -27.44 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11387090 120 - 22422827 UUUCAAUUACUCGCGUUCGCUUAAAGUGAACGCACACCAUGAAAAUAACUCUGCUGCUGGGGCCUGGUAUUUCUUGCCAGUGUCUCUGGUCACUCUGUUGCCGGACGUUUAACGCUAGGC (((((.......((((((((.....))))))))......)))))..(((...(..((..((((((((((.....)))))).))).)..))..)...)))(((((.((.....)))).))) ( -36.12, z-score = -1.79, R) >droEre2.scaffold_4690 14978930 120 + 18748788 UUUCAAUUACUCGCGUUCGCGUAAAGUGAACGCACACCAUGAAAAUAACUCUGUUGCUCUUGCCUGGUAUUUCUUGACGGCGCCUCUGGUCACGCUGUUGCCAGCCGCUUGACGAUUGGC (((((.......((((((((.....))))))))......))))).....((.((((.((..(((((((.......(((((((..........))))))))))))..))..)))))).)). ( -32.63, z-score = -0.39, R) >droYak2.chrX 11001557 120 + 21770863 UUUCAAUUACUCGCGUUCGCGUAAAGUGAACGCACACCAUGAAAAUAACUCUGUUGCUCUUGCCUGGUAUUUUUUGAAAGUGCCUCUGGUCACGCUGUGACCAGCCGUAUGACGCUUGGG (((((.......((((((((.....))))))))......)))))....(((....((((.(((..(((((((.....))))))).((((((((...))))))))..))).)).))..))) ( -36.72, z-score = -2.35, R) >droSec1.super_36 108902 120 - 449511 UUUCAAUUACUCGCGUUCGCUUAAAGUGAACGCACACCACGAAAAUAAAUCUGCUGCUGAGGCCUGGUAUUUCUUGCCAGUGCCUCUGGUCACUCUGUUGUCCUGCGUUUAACGCUAGGC ...((((.....((((((((.....))))))))..((((.((.......)).......(((((((((((.....)))))).)))))))))......)))).((((((.....))).))). ( -39.60, z-score = -3.49, R) >droSim1.chrX 8789111 120 - 17042790 UUUCAAUUACUCGCGUUCGCGUAAAGUGAACGCACACCACGAAAAUAACUUUGCUGCUGAGGCCUGGUAUUUCUUGCCAGUGCCUCUGGUCACUCUGUUGUCCCGCGCUUAACGCUAGGC ............((((((((.....))))))))...((......(((((...(..((((((((((((((.....)))))).))))).)))..)...)))))...(((.....)))..)). ( -40.70, z-score = -2.99, R) >consensus UUUCAAUUACUCGCGUUCGCGUAAAGUGAACGCACACCAUGAAAAUAACUCUGCUGCUGAGGCCUGGUAUUUCUUGCCAGUGCCUCUGGUCACUCUGUUGCCAGCCGUUUAACGCUAGGC ............((((((((.....))))))))...((.................((((((((((((((.....)))))).))))).)))...((((....))))............)). (-25.88 = -27.44 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:21 2011