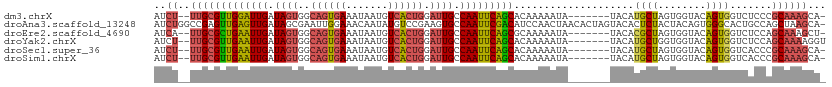
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,341,288 – 11,341,390 |
| Length | 102 |
| Max. P | 0.677396 |

| Location | 11,341,288 – 11,341,390 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Shannon entropy | 0.35473 |
| G+C content | 0.42395 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -12.85 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.677396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
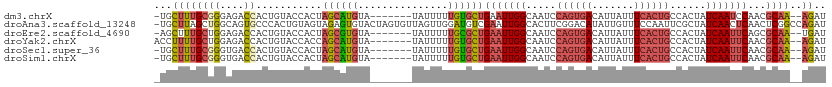
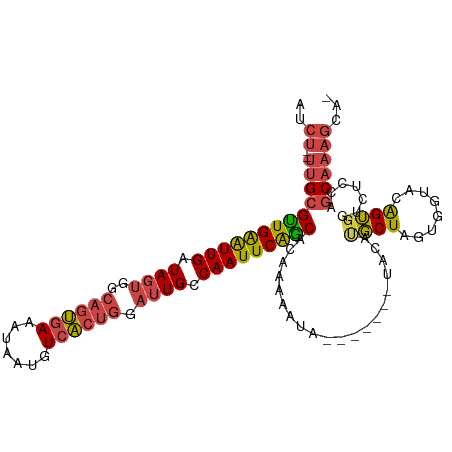
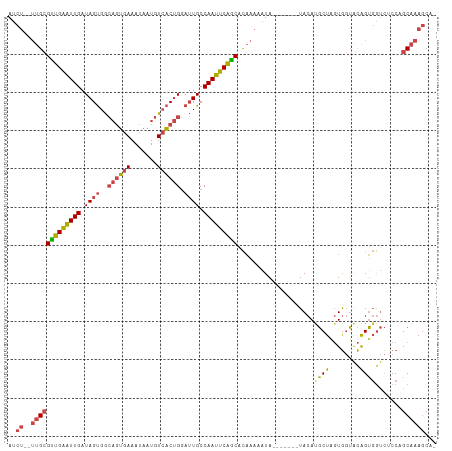
>dm3.chrX 11341288 102 + 22422827 -UGCUUUGCGGGAGACCACUGUACCACUAGCAUGUA-------UAUUUUUGUGCUGAAUUGGCAAUCCAGUGACAUUAUUUCACUGCCACUAUCAAUCCAACGCAA--AGAU -..((((((((....))...((.(((.(((((((..-------......)))))))...))).....((((((.......))))))..............))))))--)).. ( -26.30, z-score = -1.73, R) >droAna3.scaffold_13248 1812534 111 + 4840945 -UGCUUAGCUGGCAGUGCCCACUGUAGUAGAGUGUACUAGUGUUAGUUGGAUGUCGAAUUGGCACUUCGGACAUAUUGUUUCCAAUUCGCUAUCAACUCAACUCGGCCAGAU -.......((((((((((.(.(((...))).).))))).(.((((((((.(((.((((((((......((((.....)))))))))))).)))))))).))).).))))).. ( -31.70, z-score = -0.79, R) >droEre2.scaffold_4690 14934239 102 - 18748788 -AGCUUUGCUGGAGACCACUGUACCACUAGCGUGUA-------UAUUUUUGCGCUGAAUUGGCAAUCCAGUGACAUUAUUUCACUGCCACUAUCAAUUCAGCGCAA--UGAU -.((..((((((..((....))....)))))).)).-------.(((.((((((((((((((.....((((((.......))))))......))))))))))))))--.))) ( -32.90, z-score = -3.44, R) >droYak2.chrX 10959235 103 - 21770863 ACCUUUUGCUGGAGACCACUGUACCACCAGCAUGUA-------UAUUUUUGUGCUGAAUUGGCAAUCCAGUGACAUUAUUUCACUGCCACUAUCAAUUCAACGCAA--AGAU ......((((((..((....))....))))))....-------.(((((((((.((((((((.....((((((.......))))))......)))))))).)))))--)))) ( -29.90, z-score = -3.76, R) >droSec1.super_36 63268 102 + 449511 -UGCUUUGCGGGUGACCACUGUACCACUAGCAUGUA-------UAUUUUUGUGCUGAAUUGGCAAUCCAGUGACAUUAUUUCACUGCCACUAUCAAUUCAACGCAA--AGAU -..((((((((....))...((......((((((..-------......))))))(((((((.....((((((.......))))))......))))))).))))))--)).. ( -29.60, z-score = -2.61, R) >droSim1.chrX 8750106 102 + 17042790 -UGCUUUGCGGGUGACCACUGUACCACUAGCAUGUA-------UAUUUUUGUGCUGAAUUGGCAAUCCAGUGACAUUAUUUCACUGCCACUAUCAAUUCAACGCAA--AGAU -..((((((((....))...((......((((((..-------......))))))(((((((.....((((((.......))))))......))))))).))))))--)).. ( -29.60, z-score = -2.61, R) >consensus _UGCUUUGCGGGAGACCACUGUACCACUAGCAUGUA_______UAUUUUUGUGCUGAAUUGGCAAUCCAGUGACAUUAUUUCACUGCCACUAUCAAUUCAACGCAA__AGAU ......(((.((..((....))....)).))).................((((.((((((((.....((((((.......))))))......)))))))).))))....... (-12.85 = -14.27 + 1.42)
| Location | 11,341,288 – 11,341,390 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Shannon entropy | 0.35473 |
| G+C content | 0.42395 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
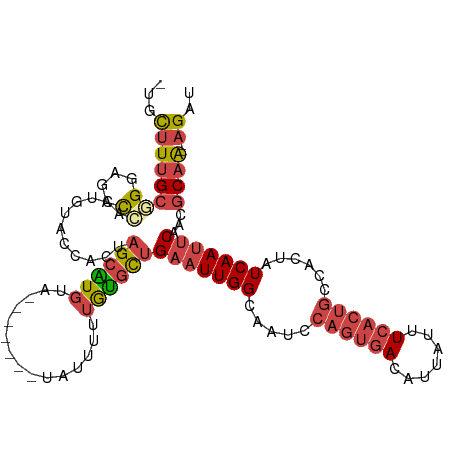
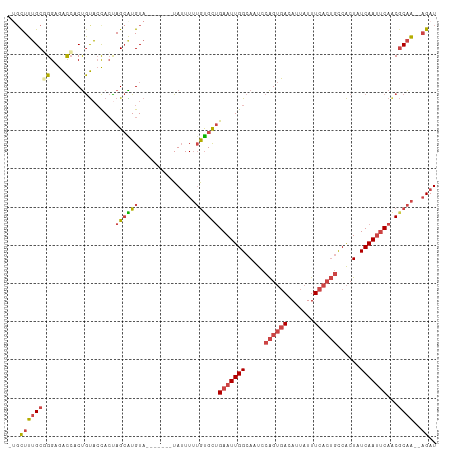
>dm3.chrX 11341288 102 - 22422827 AUCU--UUGCGUUGGAUUGAUAGUGGCAGUGAAAUAAUGUCACUGGAUUGCCAAUUCAGCACAAAAAUA-------UACAUGCUAGUGGUACAGUGGUCUCCCGCAAAGCA- ..((--(((((..(((((..((((((((.........))))))))...(((((....((((........-------....))))..)))))....)))))..)))))))..- ( -29.50, z-score = -1.39, R) >droAna3.scaffold_13248 1812534 111 - 4840945 AUCUGGCCGAGUUGAGUUGAUAGCGAAUUGGAAACAAUAUGUCCGAAGUGCCAAUUCGACAUCCAACUAACACUAGUACACUCUACUACAGUGGGCACUGCCAGCUAAGCA- ..(((((.(.(((((((((...(((..(((((.........)))))..))))))))))))..........((((((((.....))))..))))....).))))).......- ( -29.20, z-score = -1.10, R) >droEre2.scaffold_4690 14934239 102 + 18748788 AUCA--UUGCGCUGAAUUGAUAGUGGCAGUGAAAUAAUGUCACUGGAUUGCCAAUUCAGCGCAAAAAUA-------UACACGCUAGUGGUACAGUGGUCUCCAGCAAAGCU- ....--(((((((((((((.((((..((((((.......)))))).)))).))))))))))))).....-------.....(((.(..(.((....)))..))))......- ( -32.60, z-score = -2.35, R) >droYak2.chrX 10959235 103 + 21770863 AUCU--UUGCGUUGAAUUGAUAGUGGCAGUGAAAUAAUGUCACUGGAUUGCCAAUUCAGCACAAAAAUA-------UACAUGCUGGUGGUACAGUGGUCUCCAGCAAAAGGU ...(--(((.(((((((((.((((..((((((.......)))))).)))).))))))))).))))....-------....((((((.((........)).))))))...... ( -31.70, z-score = -2.35, R) >droSec1.super_36 63268 102 - 449511 AUCU--UUGCGUUGAAUUGAUAGUGGCAGUGAAAUAAUGUCACUGGAUUGCCAAUUCAGCACAAAAAUA-------UACAUGCUAGUGGUACAGUGGUCACCCGCAAAGCA- ..((--(((((((((((((.((((..((((((.......)))))).)))).))))))))).........-------.........(.(((((....)).))))))))))..- ( -28.50, z-score = -1.28, R) >droSim1.chrX 8750106 102 - 17042790 AUCU--UUGCGUUGAAUUGAUAGUGGCAGUGAAAUAAUGUCACUGGAUUGCCAAUUCAGCACAAAAAUA-------UACAUGCUAGUGGUACAGUGGUCACCCGCAAAGCA- ..((--(((((((((((((.((((..((((((.......)))))).)))).))))))))).........-------.........(.(((((....)).))))))))))..- ( -28.50, z-score = -1.28, R) >consensus AUCU__UUGCGUUGAAUUGAUAGUGGCAGUGAAAUAAUGUCACUGGAUUGCCAAUUCAGCACAAAAAUA_______UACAUGCUAGUGGUACAGUGGUCUCCAGCAAAGCA_ ......(((((((((((((.((((..((((((.......)))))).)))).)))))))))....................((((........)))).......))))..... (-18.73 = -18.90 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:11 2011