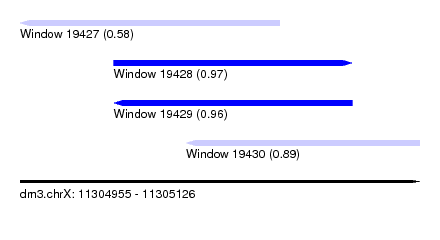
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,304,955 – 11,305,126 |
| Length | 171 |
| Max. P | 0.967501 |

| Location | 11,304,955 – 11,305,066 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Shannon entropy | 0.42148 |
| G+C content | 0.53925 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -25.54 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11304955 111 - 22422827 AGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCAC------CACCUUCGGGGGAUUCUCGUGACCGAAAUGGACCAGGGAAAUGGCCUUCUU ((((...(((((((((..(((((.(((((((.(.(((((((((...........))))).------..((....)))))).)....))))))).)))))...)))..)))))))))) ( -39.90, z-score = -1.16, R) >droSim1.chrX_random 3170745 108 - 5698898 AGGAUGCAGGCCAUCUAUGUCCAUCUUGGUCUGCAUCCUGGAGAUCCGUACAUCCUUCAC---------CUUGCAGGGCUCCUCGUGACCGAAAUGGACCAGGGAAAUGGCCUUCUU ((((...(((((((((..((((((.((((((.((.....((((.((((((..........---------..))).)))))))..)))))))).))))))...)))..)))))))))) ( -37.40, z-score = -0.98, R) >droSec1.super_36 28068 108 - 449511 AGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCAC---------CUUGGAGGGCUCCUCGUAACCGAAAUGGACCAGGGAAAUGGCCUUCUU (((((((((((((.((......))..)))))))))))))((((.(((((.((........---------..)))))))))))(((....)))...((.(((......)))))..... ( -39.20, z-score = -1.36, R) >droYak2.chrX 10922790 117 + 21770863 AGGAUGCAGGCCAUCGAUGUCCAGUUUGGUCUGCAUCCUGGAAAUCCUCACAUCCCUCACAUCCCUCACCUUCGAGUGCUCCUCGUGGCCGAAAUGGACCAGGGAAAUGGCCCUCUU (((((((((((((.(........)..)))))))))))))((....))(((..(((((....(((.((.((..((((.....)))).))..))...)))..)))))..)))....... ( -39.10, z-score = -1.02, R) >droEre2.scaffold_4690 14898583 108 + 18748788 AGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUCACAUCCUUCACA---------UUGGCCUGCUCCUCGUGGCUGAAAUGGACCAGGGUGAUGGCCUUCUU (((((((((((((.((......))..)))))))))))))((((..(.((((..((......---------..))((((.(((.............))).)))))))).)..)))).. ( -42.62, z-score = -1.89, R) >droAna3.scaffold_13248 1774529 108 - 4840945 CGGACGCAGGCCAUUCCACUCCAGCUUGUUGUUCAUUUUGGAGACCCUCAGGUCCCUGAA---------CUUGGUGGGUGCUCCCUUGUCGGCCAUUCCAUUGAUAAUCGCCCGUCU .(((((..((((......((((((..((.....))..))))))((((((((((......)---------))))).))))...........))))........(.....)...))))) ( -32.20, z-score = -0.28, R) >consensus AGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUGGAGAUCCUCACAUCCUUCAC_________CUUGGAGGGCUCCUCGUGACCGAAAUGGACCAGGGAAAUGGCCUUCUU (((((((((((((.............)))))))))))))(((..........)))..................(((((((.(((....((.....))....)))....))))))).. (-25.54 = -26.10 + 0.56)

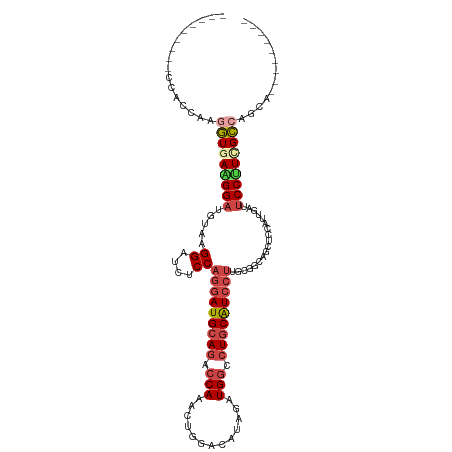
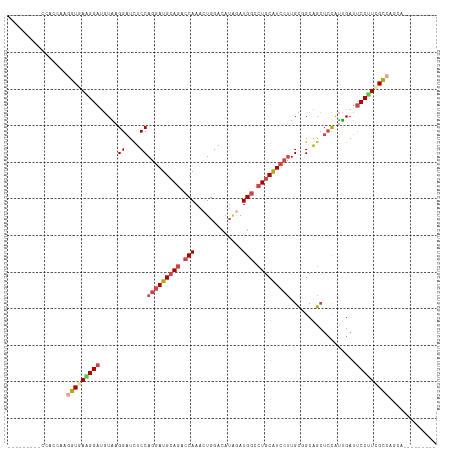
| Location | 11,304,995 – 11,305,097 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.28 |
| Shannon entropy | 0.46595 |
| G+C content | 0.54250 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -24.54 |
| Energy contribution | -25.74 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11304995 102 + 22422827 ------CCCCGAAGGUGGUGAAGGAUGUAAGGAUCUCCAGGAUGCAGACCAAACUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCCAUUGAUUCCUUUGCCAACA--------- ------.......(.(((..(((((..((((((.(((((((((((((.(((..(((....))).))).)))))))))...)).)).))).)))..)))))..))).).--------- ( -40.50, z-score = -2.93, R) >droSim1.chrX_random 3170785 99 + 5698898 ---------CCUGCAAGGUGAAGGAUGUACGGAUCUCCAGGAUGCAGACCAAGAUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCUAUUGAUUCCUUCGCCAGCA--------- ---------...((..(((((((((.....(((.(((((((((((((.(((..((....))...))).)))))))))...)).)).)))......))))))))).)).--------- ( -38.60, z-score = -2.32, R) >droSec1.super_36 28108 99 + 449511 ---------CCUCCAAGGUGAAGGAUGUAAGGAUCUCCAGGAUGCAGACCAAGCUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCCAUUGAUUCCUUCGCCAGCA--------- ---------.......(((((((((..((((((.(((((((((((((.(((..(((....))).))).)))))))))...)).)).))).)))..)))))))))....--------- ( -42.70, z-score = -3.22, R) >droYak2.chrX 10922830 108 - 21770863 ACUCGAAGGUGAGGGAUGUGAGGGAUGUGAGGAUUUCCAGGAUGCAGACCAAACUGGACAUCGAUGGCCUGCAUCCUUGCUGCGGCUCCGUUGAUUCCCUGGCCAGUG--------- .......(((.(((((..(((.(((.((..((....))(((((((((.(((.............))).))))))))).......))))).)))..))))).)))....--------- ( -40.22, z-score = -1.06, R) >droEre2.scaffold_4690 14898623 99 - 18748788 ---------AGGCCAAUGUGAAGGAUGUGAGGAUCUCCAGGAUGCAGACCAAACUGGACAUAGAUGGCCUGCAUCCUUGCUGCAGCUCCAUUGAUUCCCUUGCCAUCA--------- ---------.(((.((.(.(((.((((.(((...(..((((((((((.(((..(((....))).))).)))))))).))..)...)))))))..)))).)))))....--------- ( -30.50, z-score = -0.08, R) >droAna3.scaffold_13248 1774569 108 + 4840945 ---------CCACCAAGUUCAGGGACCUGAGGGUCUCCAAAAUGAACAACAAGCUGGAGUGGAAUGGCCUGCGUCCGUGUCGCAAGAACACCGAUCCCUUCGGCGUGGAUCGCAUCA ---------((((...(((((((((((....)))))).....))))).....(((((((.(((..(((....))).(((((....).))))...))))))))))))))......... ( -38.70, z-score = -1.35, R) >consensus _________CCACCAAGGUGAAGGAUGUAAGGAUCUCCAGGAUGCAGACCAAACUGGACAUAGAUGGCCUGCAUCCUUGCGGCAGCUCCAUUGAUUCCUUCGCCAGCA_________ ................(((((((((.....((....))(((((((((.(((.............))).)))))))))..................)))))))))............. (-24.54 = -25.74 + 1.20)
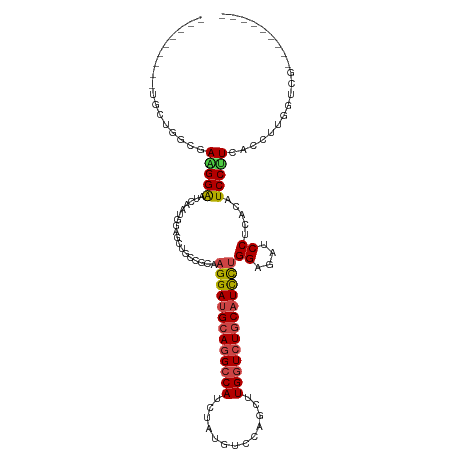
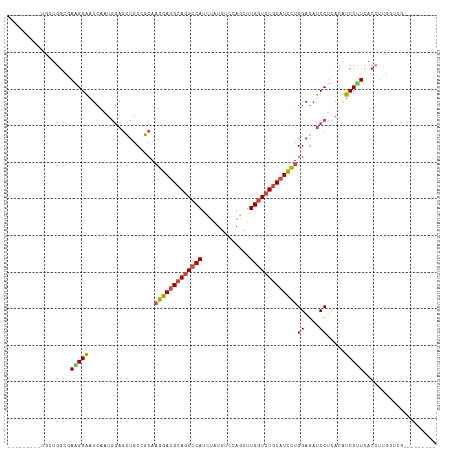
| Location | 11,304,995 – 11,305,097 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.28 |
| Shannon entropy | 0.46595 |
| G+C content | 0.54250 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11304995 102 - 22422827 ---------UGUUGGCAAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCACCACCUUCGGGG------ ---------...(((..(((((......(((.((.((...(((((((((((((.((......))..))))))))))))))))).))).....)))))..)))((....)).------ ( -42.00, z-score = -3.04, R) >droSim1.chrX_random 3170785 99 - 5698898 ---------UGCUGGCGAAGGAAUCAAUAGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAUCUUGGUCUGCAUCCUGGAGAUCCGUACAUCCUUCACCUUGCAGG--------- ---------(((.((.((((((.........((....)).(((((((((((((.............)))))))))))))((....)).....)))))).))..)))..--------- ( -40.42, z-score = -3.00, R) >droSec1.super_36 28108 99 - 449511 ---------UGCUGGCGAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUGGAGAUCCUUACAUCCUUCACCUUGGAGG--------- ---------....((.((((((......(((.((.((...(((((((((((((.((......))..))))))))))))))))).))).....)))))).)).......--------- ( -40.70, z-score = -2.28, R) >droYak2.chrX 10922830 108 + 21770863 ---------CACUGGCCAGGGAAUCAACGGAGCCGCAGCAAGGAUGCAGGCCAUCGAUGUCCAGUUUGGUCUGCAUCCUGGAAAUCCUCACAUCCCUCACAUCCCUCACCUUCGAGU ---------....((..(((((......(((((....)).(((((((((((((.(........)..)))))))))))))((....)).....)))......)))))..))....... ( -36.40, z-score = -1.44, R) >droEre2.scaffold_4690 14898623 99 + 18748788 ---------UGAUGGCAAGGGAAUCAAUGGAGCUGCAGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUGGAGAUCCUCACAUCCUUCACAUUGGCCU--------- ---------.((((...(((((......(((.((.((....((((((((((((.((......))..)))))))))))))).)).))).....)))))..)))).....--------- ( -34.90, z-score = -0.68, R) >droAna3.scaffold_13248 1774569 108 - 4840945 UGAUGCGAUCCACGCCGAAGGGAUCGGUGUUCUUGCGACACGGACGCAGGCCAUUCCACUCCAGCUUGUUGUUCAUUUUGGAGACCCUCAGGUCCCUGAACUUGGUGG--------- ............(((((((((((((((....((((((.......)))))).....)).((((((..((.....))..)))))).......)))))))....)))))).--------- ( -36.00, z-score = -0.41, R) >consensus _________UGCUGGCGAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUGGAGAUCCUCACAUCCUUCACCUUGGUCG_________ .................(((((..................(((((((((((((.............)))))))))))))((....)).....))))).................... (-23.75 = -24.05 + 0.31)

| Location | 11,305,026 – 11,305,126 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 70.53 |
| Shannon entropy | 0.51028 |
| G+C content | 0.55056 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -23.25 |
| Energy contribution | -24.54 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11305026 100 - 22422827 ---------------------AUAGGCUUUGGAUGACUGCGGGCUGUCAGUGUUGGCAAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUG ---------------------...((((((.(((..((((.(((.......))).))..))..)))...))))))......(((((((((((((.((......))..))))))))))))). ( -37.80, z-score = -1.38, R) >droSim1.chrX_random 3170813 100 - 5698898 ---------------------GUAGGCUUUCUUGGACUGCGGGUUGUCAGUGCUGGCGAAGGAAUCAAUAGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAUCUUGGUCUGCAUCCUG ---------------------...(((.((((..((.(.(...((((((....)))))).).).))...))))..)))...(((((((((((((.............))))))))))))). ( -38.12, z-score = -1.57, R) >droSec1.super_36 28136 100 - 449511 ---------------------GUAGGCUUUGGAGGACGGCGGGUUGUCAGUGCUGGCGAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUG ---------------------.............(.((((((.((((((....))))((.....))....)).))))))).(((((((((((((.((......))..))))))))))))). ( -38.10, z-score = -0.68, R) >droYak2.chrX 10922867 100 + 21770863 ---------------------GUAGGCUUGGUAGGACUUUGGUUUCUUGCCACUGGCCAGGGAAUCAACGGAGCCGCAGCAAGGAUGCAGGCCAUCGAUGUCCAGUUUGGUCUGCAUCCUG ---------------------...(((((.((.......((((((((((((...)).)))))))))))).)))))......(((((((((((((.(........)..))))))))))))). ( -43.41, z-score = -2.80, R) >droEre2.scaffold_4690 14898651 100 + 18748788 ---------------------GUAGGCUUGGGAGGACUGCGGCUUGCCACUGAUGGCAAGGGAAUCAAUGGAGCUGCAGCAAGGAUGCAGGCCAUCUAUGUCCAGUUUGGUCUGCAUCCUG ---------------------.............(.((((((((..(((.((((.........)))).)))))))))))).(((((((((((((.((......))..))))))))))))). ( -42.40, z-score = -1.94, R) >droAna3.scaffold_13248 1774597 120 - 4840945 AUCAGCAUGUGAUACAUCCGGGCACGGUCUACCGUAUCAUUGA-UGCGAUCCACGCCGAAGGGAUCGGUGUUCUUGCGACACGGACGCAGGCCAUUCCACUCCAGCUUGUUGUUCAUUUUG ....(((((((((((..(((....)))......)))))))..)-)))(((((.(......)))))).(((((.....)))))((((((((((............))))).)))))...... ( -35.20, z-score = 0.07, R) >consensus _____________________GUAGGCUUUGGAGGACUGCGGGUUGUCAGUGCUGGCGAAGGAAUCAAUGGAGCUGCCGCAAGGAUGCAGGCCAUCUAUGUCCAGCUUGGUCUGCAUCCUG ........................(((((((...((.(.(...((((((....)))))).).).))..)))))))......(((((((((((((.............))))))))))))). (-23.25 = -24.54 + 1.29)
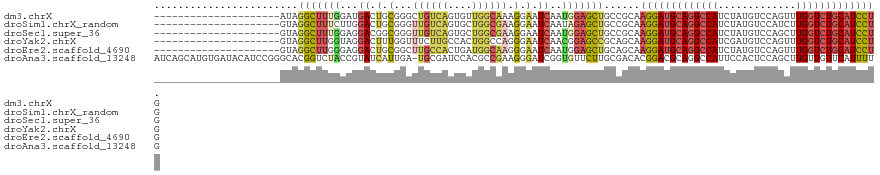

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:07 2011