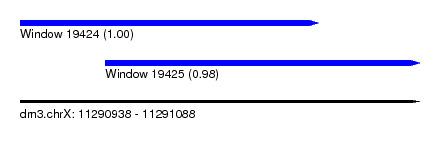
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,290,938 – 11,291,088 |
| Length | 150 |
| Max. P | 0.999044 |

| Location | 11,290,938 – 11,291,050 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.81 |
| Shannon entropy | 0.51204 |
| G+C content | 0.36388 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -12.87 |
| Energy contribution | -14.15 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
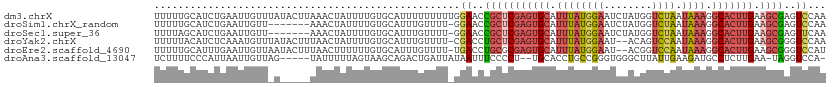
>dm3.chrX 11290938 112 + 22422827 UUUUUGCAUCUGAAUUGUUUAUACUUAAACUAUUUUGUGCAUUUUUUUUUGGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUCCAA ....(((((..((((.(((((....))))).)))).))))).......(((((..(((((((((((.((((((((........))).))))).))))))).))))..))))) ( -35.80, z-score = -5.03, R) >droSim1.chrX_random 3156828 104 + 5698898 UUUUUGCAUCUGAAUUGUU-------AAACUAUUUUGUGCAUUUGUUUU-GGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUCCAA ....(((((..((((.(..-------...).)))).)))))......((-(((..(((((((((((.((((((((........))).))))).))))))).))))..))))) ( -31.30, z-score = -3.42, R) >droSec1.super_36 14201 104 + 449511 UUUUAGCAUCUGAAUUGUU-------AAACUAUUUUGUGCAUUUGUUUU-GGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUUCAA .....((((..((((.(..-------...).)))).)))).........-.(((((((((((((((.((((((((........))).))))).))))))).)))).)))).. ( -29.70, z-score = -2.93, R) >droYak2.chrX 10908536 109 - 21770863 UUUUUACAUCUCAAAUGUUUAUACUUUAACUAUUUUGUGCAUUUGUUUU-CGACCUGCUCGAGUGCAUUUAUGGAAU--ACAGUCCAAUAAAGGCACUUGAAGCGGGUCCAA ...........(((((((....................)))))))....-.(((((((((((((((.((((((((..--....))).))))).))))))).))))))))... ( -34.95, z-score = -5.17, R) >droEre2.scaffold_4690 14884992 109 - 18748788 UUUUUGCAUUUGAAUUGUUAAUACUUUAACUUUUUUGUGCAUUUGUUUU-UGACCUGCGCGAGUGCAUUUAUGGAAU--ACGGUCCAAUAAAGGCACUUGAAGCGGGUCCAU ....(((((..(((..(((((....)))))..))).)))))........-.(((((((.(((((((.((((((((..--....))).))))).)))))))..)))))))... ( -37.90, z-score = -5.04, R) >droAna3.scaffold_13047 630498 103 - 1816235 UCUUUUCCCAUUAAUUGUUAG-----UAUUUUUAGUAAGCAGACUGAUUAUAAUUUCCCCU--UGCACCUGCCGGGUGGGCUUAUUGAAGAUGCCUCUUGAA-UAGGUCCA- ......(((............-----(((..(((((......)))))..))).........--.((....)).)))((((((((((.((((....)))).))-))))))))- ( -24.60, z-score = -1.07, R) >consensus UUUUUGCAUCUGAAUUGUU_A_____AAACUAUUUUGUGCAUUUGUUUU_GGAACCGCUCGAGUGCAUUUAUGGAAUCUACGGUCUAAUAAAGGCACUUGAAGCGAGUCCAA ...................................................((..(((((((((((.((((((((........)))).)))).))))))).))))..))... (-12.87 = -14.15 + 1.28)
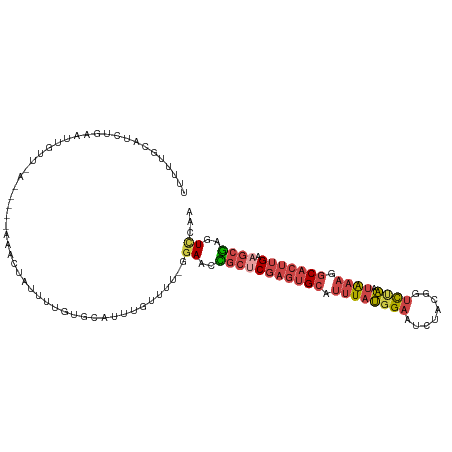
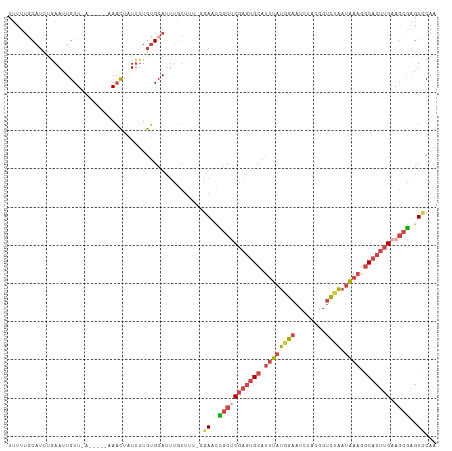
| Location | 11,290,970 – 11,291,088 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Shannon entropy | 0.27267 |
| G+C content | 0.41721 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.58 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11290970 118 + 22422827 UUUUGUGCAUUUUUUUUUGGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUCCAAUUCCUUACU--CCAAUCAUCCCAAUUCUUCCUCGUGUACA ...(((((((......(((((..(((((((((((.((((((((........))).))))).))))))).))))..))))).........--..(((.......))).......))))))) ( -29.40, z-score = -2.62, R) >droSim1.chrX_random 3156853 111 + 5698898 UUUUGUGCAUUUGUUUU-GGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUCCAAUUCCUUACUCUCCAAUCAUCCCAAUCCUUGCU-------- ......(((...(..((-((....((((((((((.((((((((........))).))))).))))))).)))((((..........))))..........))))..).))).-------- ( -27.80, z-score = -2.07, R) >droSec1.super_36 14226 111 + 449511 UUUUGUGCAUUUGUUUU-GGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUUCAAUUCCUUACUCUCCAAUCAUCCCAAUCCUUCCU-------- ...........((..((-(((...((((((((((.((((((((........))).))))).))))))).)))((((..........))))))))).))..............-------- ( -25.50, z-score = -1.71, R) >droYak2.chrX 10908568 106 - 21770863 UUUUGUGCAUUUGUUUU-CGACCUGCUCGAGUGCAUUUAUGGAAU--ACAGUCCAAUAAAGGCACUUGAAGCGGGUCCAAUUCCUUACU--CCAAUCCUUCCCUAAGUACA--------- ...(((((....(....-)(((((((((((((((.((((((((..--....))).))))).))))))).))))))))............--...............)))))--------- ( -33.50, z-score = -4.42, R) >droEre2.scaffold_4690 14885024 106 - 18748788 UUUUGUGCAUUUGUUUU-UGACCUGCGCGAGUGCAUUUAUGGAAU--ACGGUCCAAUAAAGGCACUUGAAGCGGGUCCAUUUCCUCACU--CCAAUCCUUCCUCAUGUACA--------- ...(((((((..(....-.(((((((.(((((((.((((((((..--....))).))))).)))))))..)))))))............--..........)..)))))))--------- ( -30.53, z-score = -2.44, R) >consensus UUUUGUGCAUUUGUUUU_GGAACCGCUCGAGUGCAUUUAUGGAAUCUAUGGUCUAAUAAAGGCACUUGAAGCGAGUCCAAUUCCUUACU__CCAAUCAUCCCAAUCCUUCCU________ ..................(((..(((((((((((.((((((((........))).))))).))))))).))))..))).......................................... (-21.62 = -21.58 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:03 2011