| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,290,638 – 11,290,784 |
| Length | 146 |
| Max. P | 0.994082 |

| Location | 11,290,638 – 11,290,784 |
|---|---|
| Length | 146 |
| Sequences | 6 |
| Columns | 163 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Shannon entropy | 0.44328 |
| G+C content | 0.40586 |
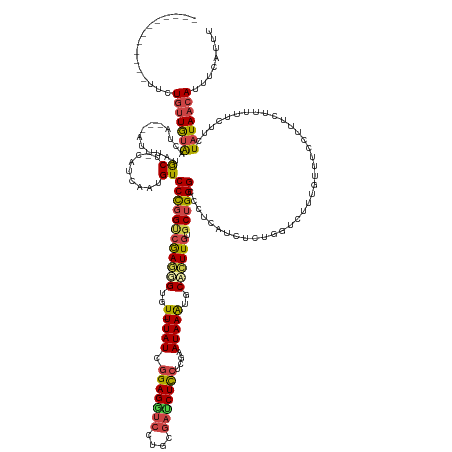
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
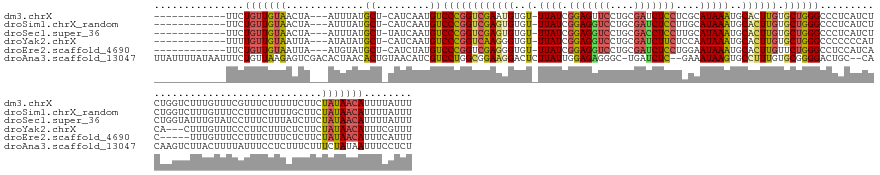
>dm3.chrX 11290638 146 + 22422827 ------------UUCUGUUGUAACUA---AUUUAUGCU-CAUCAAUGUCCCGGUCGAAUGUGU-UUAUCGGAGUUCCUGCGAUCUCCUCGCAUAAAUGCACUUGUGCUGGGCCCUCAUCUCUGGUCUUUGUUUCGUUUCUUUUUCUUCUAUAACAUUUUAUUU ------------...(((((((....---.........-.........(((((((((.(((((-((((.((....)).((((.....))))))))))))).))).)))))).((........))........................)))))))........ ( -28.40, z-score = -2.11, R) >droSim1.chrX_random 3156528 146 + 5698898 ------------UUCUGUUGUAACUA---AUUUAUGCU-CAUCAAUGUCCCGGUCGAGUGUGU-UUAUCGGAGGUCCUGCGAUCUCCUUGCAUAAAUGCACUUGUGCUGGGCCCUCAUCUCUGGUCUUUGUUUCCUUUCUUUUGCUUCUAUAACAUUUUAUUU ------------...(((((((....---.........-.........(((((((((((((((-((((.(((((((....)))))))....))))))))))))).)))))).((........))........................)))))))........ ( -37.60, z-score = -4.08, R) >droSec1.super_36 13901 146 + 449511 ------------UUCUGUUGUAACUA---AUUUAUGCU-UAUCAAUGUCCCGGUCGAGUGUGU-UUAUCGGAGGUCCUGCGACCUCCUUGCAUAAAUGCACUUGUGCUGGGCCCUCAUCUCUGGUAUUUGUAUCCUUUCUUUAUCUUCUAUAACAUUUUAUUU ------------...(((((((....---....((((.-((((((((.(((((((((((((((-((((.(((((((....)))))))....))))))))))))).))))))....)))...)))))...))))...............)))))))........ ( -40.80, z-score = -5.18, R) >droYak2.chrX 10908205 143 - 21770863 ------------UUUUGUUGUAAUUA---AUAUAUGCU-CAUCAAUGUCCCGGUCAAGGGUGU-UUAUCGGAGGUCCUGCGAUCUUCUCCAAUAAAUGCACUUGUGCUGGGCCCCCCCAUCA---CUUUGUUUCCCUUCUUUCUCUUCUAUAACAUUUCGUUU ------------...(((((((....---..((((...-.....))))((((((((((.((((-((((.(((((...........))))).)))))))).)))).))))))...........---.......................)))))))........ ( -30.10, z-score = -1.74, R) >droEre2.scaffold_4690 14884660 141 - 18748788 ------------UUCUGUUGUAAUUA---AUGUAUGCU-CAUCUAUGUCCCGGUCGAGGGUGU-UUAUCGGAGGUCCUGCGAUCUCCUGGAAUAAAUGCACUUGUUCUGGGCCUCCAUCAC-----UUUGUUUCCUUUCUUUCUCUUCUAUAACAUUUCAUUU ------------...(((((((((..---(((......-)))..))(.(((((.((((.((((-((((.(((((((....)))))))....)))))))).))))..))))).)........-----......................)))))))........ ( -30.60, z-score = -1.34, R) >droAna3.scaffold_13047 630340 158 - 1816235 UUAUUUUAUAAUUUCUGUUAAGAGUCGACACUAACACUGUAACAUCGUCCUGGCGGAAGGACUCUUAUUGGAGAGGGC-UGAUCUC--GAAAUAAGUGCCUUUGUGCGGGGACUGC--CACAAGUCUUACUUUUAUUUCCUCUUUCUUUCUAUAAUUUCCUCU ..................(((((((.(((((.......))......(((((.((((((((.(.((((((.((((....-...))))--..)))))).)))))).))).)))))...--.....)))..)))))))............................ ( -34.70, z-score = -0.01, R) >consensus ____________UUCUGUUGUAACUA___AUUUAUGCU_CAUCAAUGUCCCGGUCGAGGGUGU_UUAUCGGAGGUCCUGCGAUCUCCUCGAAUAAAUGCACUUGUGCUGGGCCCUCAUCUCUGGUCUUUGUUUCCUUUCUUUUUCUUCUAUAACAUUUCAUUU ...............(((((((.............((.........))((((((((((((..(.((((.(((((((....)))))))....)))))..)))))).)))))).....................................)))))))........ (-22.64 = -23.12 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:34:02 2011