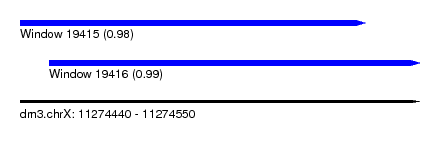
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,274,440 – 11,274,550 |
| Length | 110 |
| Max. P | 0.988051 |

| Location | 11,274,440 – 11,274,535 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Shannon entropy | 0.27924 |
| G+C content | 0.54633 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -25.20 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
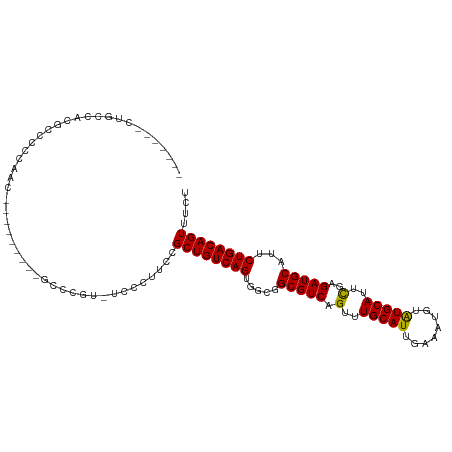
>dm3.chrX 11274440 95 + 22422827 -------CAGCCACACCCCCAAC---------GCCCAU-UCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCU -------.............(((---------......-........(((((.....)))))(((((..((((((..((((....))))...))))))..))))).)))... ( -26.70, z-score = -1.89, R) >droSim1.chrX 8704931 95 + 17042790 -------CUGCCACGCCCCCAAC---------GCCCGU-UCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCU -------(((((((((.......---------)).((.-.......))......))))))).(((((..((((((..((((....))))...))))))..)))))....... ( -29.10, z-score = -2.06, R) >droSec1.super_15 1953111 95 + 1954846 -------CUGCCACGCCUCCAAC---------GCCCGU-UCCCUUCCGCUGUCAGUGGCAGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCU -------(((((((((.......---------)).((.-.......))......))))))).(((((..((((((..((((....))))...))))))..)))))....... ( -29.80, z-score = -2.59, R) >droYak2.chrX 10892140 104 - 21770863 -------CUGCCACGCCCCCUACCCGUUGCAAACGCGC-CCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUGUGCAUUCGAGAUGCAUUCUGACAGUUUCU -------(((((((((............))....(((.-.......))).....))))))).(((((..((((((..((((....))))...))))))..)))))....... ( -33.00, z-score = -2.02, R) >droAna3.scaffold_13047 611851 101 - 1816235 GGGGGUUCCGGUACCCGUCCGCCC--------ACCCGUACCCCCAGCGCUGUCAGUG---GCGUCAGUUUGCAUUGAAAUGUGUGCAUUUGAGAUGCAUUCUGACAGUUUCU ((((((.(.(((...(....)...--------))).).))))))...((((((((..---(((((.(..(((((........)))))..)..)))))...)))))))).... ( -40.10, z-score = -3.22, R) >consensus _______CUGCCACGCCCCCAAC_________GCCCGU_UCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCU ...............................................((((((((.....(((((.(..(((((........)))))..)..)))))...)))))))).... (-25.20 = -24.80 + -0.40)

| Location | 11,274,448 – 11,274,550 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Shannon entropy | 0.26565 |
| G+C content | 0.53673 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -25.20 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.988051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11274448 102 + 22422827 ---------CCCCCAACGCCCAU-UCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCUGAUUUCCGCUUCCCG ---------........((....-........(((((.....)))))(((((..((((((..((((....))))...))))))..)))))..............))...... ( -27.80, z-score = -2.27, R) >droSim1.chrX 8704939 102 + 17042790 ---------CCCCCAACGCCCGU-UCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCUGAUUUCCGCUUCCCG ---------........((....-........(((((.....)))))(((((..((((((..((((....))))...))))))..)))))..............))...... ( -27.80, z-score = -2.05, R) >droSec1.super_15 1953119 102 + 1954846 ---------CCUCCAACGCCCGU-UCCCUUCCGCUGUCAGUGGCAGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCUGAUUUCCGCUUCCCG ---------........((....-........(((((.....)))))(((((..((((((..((((....))))...))))))..)))))..............))...... ( -28.50, z-score = -2.68, R) >droYak2.chrX 10892148 111 - 21770863 CCCCCUACCCGUUGCAAACGCGC-CCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUGUGCAUUCGAGAUGCAUUCUGACAGUUUCUGAUUUCCGCUUCCCG ........((((..(..(((((.-.......))).))..)..)))).(((((..((((((..((((....))))...))))))..)))))...................... ( -31.80, z-score = -2.19, R) >droAna3.scaffold_13047 611866 95 - 1816235 --------CGUCCGCCCACCCGUACCCCCAGCGCUGUCAGUG---GCGUCAGUUUGCAUUGAAAUGUGUGCAUUUGAGAUGCAUUCUGACAGUUUCUGAUUUCCGC------ --------......................(((..(((((..---(((((((..((((((.(((((....)))))..))))))..))))).))..)))))...)))------ ( -30.80, z-score = -2.97, R) >consensus _________CCCCCAACGCCCGU_UCCCUUCCGCUGUCAGUGGCGGCGUCAGUUUGCAUUGAAAUGUAUGCAUUCGAGAUGCAUUCUGACAGUUUCUGAUUUCCGCUUCCCG ................................((((((((.....(((((.(..(((((........)))))..)..)))))...))))))))................... (-25.20 = -24.80 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:56 2011