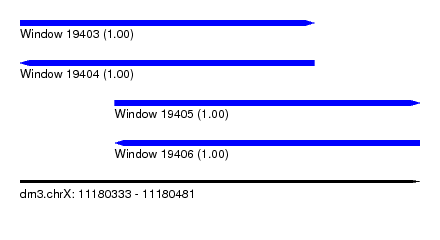
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,180,333 – 11,180,481 |
| Length | 148 |
| Max. P | 0.999901 |

| Location | 11,180,333 – 11,180,442 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Shannon entropy | 0.25192 |
| G+C content | 0.32153 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.89 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.997870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11180333 109 + 22422827 AAUAUAACCUACAAAAAAAA-----AGAAUAAAAAUUAUAGCUUGGACAACAGUACGUAUGAGUAACAUCUUAGCUCACAGAUUGCUCAUACGCACCGUUGUUUC-CACUUGCGA ....................-----...............((.((((((((.((.((((((((((..((((........)))))))))))))).)).))))..))-))...)).. ( -26.40, z-score = -3.98, R) >droYak2.chrX 10807518 113 - 21770863 AAUAUAACUUUCAAACUAAAUUACAAUAAUAAUAAUUAGAAACUGUACAACAGUACGUAUGAGUAAUA-CUUAGCAUUCAGAUUGCUCAUACGCACCGUUGUUUU-CACUGAAGA ........(((((.............((((....))))((((....(((((.((.((((((((((((.-((........)))))))))))))).)).))))))))-)..))))). ( -24.90, z-score = -3.51, R) >droSec1.super_15 1865614 110 + 1954846 AAUAUAACCUAUAACUACAA-----AUAAUAAUAAUUAUAACCUGUACAACAGUACGUAUGGGUAACAUCUUAGCUCACAAAUUGCUCAUACGCACCGUUGUUUUUCACUUGAGA ..............((.(((-----(((((....))))).......(((((.((.(((((((((((................))))))))))).)).))))).......))))). ( -18.99, z-score = -1.60, R) >droSim1.chrX_random 3137236 110 + 5698898 AAUAUAACCUAUAACUACAA-----AUAAUAAUAAUUAUAACCUGCACAACAGUGCGUAUGAGUAACAUCUUAGCUCACAAAUUGCUCAUACGCACCGUUGUUUUUCACUUCAGA ....................-----.....................(((((.((((((((((((((................)))))))))))))).)))))............. ( -25.69, z-score = -4.83, R) >consensus AAUAUAACCUACAAAUAAAA_____AUAAUAAUAAUUAUAACCUGUACAACAGUACGUAUGAGUAACAUCUUAGCUCACAAAUUGCUCAUACGCACCGUUGUUUU_CACUUGAGA ..............................................(((((.((.(((((((((((................))))))))))).)).)))))............. (-19.08 = -18.89 + -0.19)

| Location | 11,180,333 – 11,180,442 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Shannon entropy | 0.25192 |
| G+C content | 0.32153 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -27.30 |
| Energy contribution | -28.42 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.998971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11180333 109 - 22422827 UCGCAAGUG-GAAACAACGGUGCGUAUGAGCAAUCUGUGAGCUAAGAUGUUACUCAUACGUACUGUUGUCCAAGCUAUAAUUUUUAUUCU-----UUUUUUUUGUAGGUUAUAUU ..((...((-((..((((((((((((((((..((((........))))....)))))))))))))))))))).))((((((((.((....-----.......)).)))))))).. ( -35.00, z-score = -4.52, R) >droYak2.chrX 10807518 113 + 21770863 UCUUCAGUG-AAAACAACGGUGCGUAUGAGCAAUCUGAAUGCUAAG-UAUUACUCAUACGUACUGUUGUACAGUUUCUAAUUAUUAUUAUUGUAAUUUAGUUUGAAAGUUAUAUU ..(((((..-...(((((((((((((((((.(((((........))-.))).))))))))))))))))).......((((..(((((....))))))))).)))))......... ( -31.30, z-score = -3.86, R) >droSec1.super_15 1865614 110 - 1954846 UCUCAAGUGAAAAACAACGGUGCGUAUGAGCAAUUUGUGAGCUAAGAUGUUACCCAUACGUACUGUUGUACAGGUUAUAAUUAUUAUUAU-----UUGUAGUUAUAGGUUAUAUU .............(((((((((((((((((((.((((.....)))).))))...)))))))))))))))..((.((((((((((......-----..)))))))))).))..... ( -29.80, z-score = -2.85, R) >droSim1.chrX_random 3137236 110 - 5698898 UCUGAAGUGAAAAACAACGGUGCGUAUGAGCAAUUUGUGAGCUAAGAUGUUACUCAUACGCACUGUUGUGCAGGUUAUAAUUAUUAUUAU-----UUGUAGUUAUAGGUUAUAUU ((((.........(((((((((((((((((..((((........))))....))))))))))))))))).))))((((((((((......-----..))))))))))........ ( -35.80, z-score = -4.43, R) >consensus UCUCAAGUG_AAAACAACGGUGCGUAUGAGCAAUCUGUGAGCUAAGAUGUUACUCAUACGUACUGUUGUACAGGUUAUAAUUAUUAUUAU_____UUGUAGUUAUAGGUUAUAUU .............(((((((((((((((((..((((........))))....)))))))))))))))))..((.((((((((((.............)))))))))).))..... (-27.30 = -28.42 + 1.12)

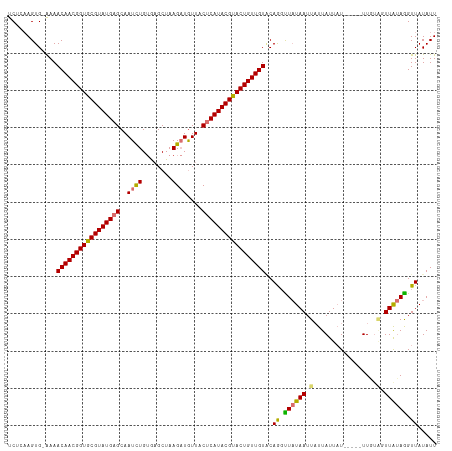
| Location | 11,180,368 – 11,180,481 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.68 |
| Shannon entropy | 0.66024 |
| G+C content | 0.37747 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -10.19 |
| Energy contribution | -12.81 |
| Covariance contribution | 2.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11180368 113 + 22422827 GCUUGGACAACAGUACGUAUGAGUAACAUCUUAGCUCACAGAUUGCUCAUACGCACCGUUGUUUC-CACUUGCGAGU------CAUUUAAACAGCUUGAAAAGCUAGGAACUUUAAAAAA (((((((((((.((.((((((((((..((((........)))))))))))))).)).))))..))-).....(((((------..........)))))..))))................ ( -29.20, z-score = -2.22, R) >droAna3.scaffold_13248 2736543 102 + 4840945 ----------UAACGCCCACACUGCGUA---UACGUAUUAUAUUACUCAUACGCACUGUUGCCUC-CAACCAGGACCUU----GUUUUAAAGUCCUUUUUAAAGUCGUCCUCUGGCGUAG ----------..(((((.(((.((((((---(..(((......)))..))))))).)))......-.....(((((...----.((((((((....))))))))..)))))..))))).. ( -27.90, z-score = -4.17, R) >droEre2.scaffold_4690 14781171 118 - 18748788 AACUGUACACCACCACGUAUGAGUUAUA-UUUAGCACGCAGAUUGCUCAUACGCACCGUUGUUUU-CACUGAAGACAUUUGCACAUUUAAGCCGUUUUAAAGGCUAGAAACUUAAAGAAA ...(((..........((((((((...(-(((.(....))))).))))))))(((....((((((-(...)))))))..))))))(((.((((........)))).)))........... ( -25.60, z-score = -1.77, R) >droYak2.chrX 10807558 112 - 21770863 AACUGUACAACAGUACGUAUGAGUAAUA-CUUAGCAUUCAGAUUGCUCAUACGCACCGUUGUUUU-CACUGAAGACU------CAUUUAAACAGCUUUAAAGGCUAGAAACUUUAAAAAA .((((.....)))).((((((((((((.-((........))))))))))))))....(((((((.-...(((....)------))...)))))))(((((((........)))))))... ( -29.30, z-score = -3.69, R) >droSec1.super_15 1865649 105 + 1954846 ACCUGUACAACAGUACGUAUGGGUAACAUCUUAGCUCACAAAUUGCUCAUACGCACCGUUGUUUUUCACUUGAGAGU------CAUUUAAAGUGUUUUAAACUUUAAAAAA--------- ......(((((.((.(((((((((((................))))))))))).)).)))))(((((....))))).------..((((((((.......))))))))...--------- ( -24.79, z-score = -2.51, R) >droSim1.chrX_random 3137271 105 + 5698898 ACCUGCACAACAGUGCGUAUGAGUAACAUCUUAGCUCACAAAUUGCUCAUACGCACCGUUGUUUUUCACUUCAGAGU------CAUUUAAAGUGCUUUAAACUUUAAAAAA--------- ......(((((.((((((((((((((................)))))))))))))).)))))...............------..((((((((.......))))))))...--------- ( -30.99, z-score = -5.06, R) >consensus ACCUGUACAACAGUACGUAUGAGUAACA_CUUAGCUCACAGAUUGCUCAUACGCACCGUUGUUUU_CACUUAAGACU______CAUUUAAACUGCUUUAAAAGCUAGAAACUU_AA_AAA ......(((((.((.(((((((((((................))))))))))).)).))))).......................................................... (-10.19 = -12.81 + 2.61)

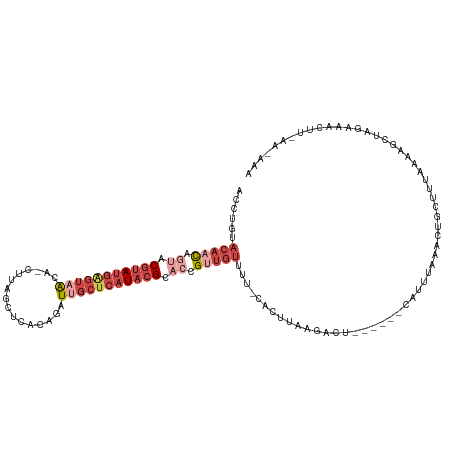
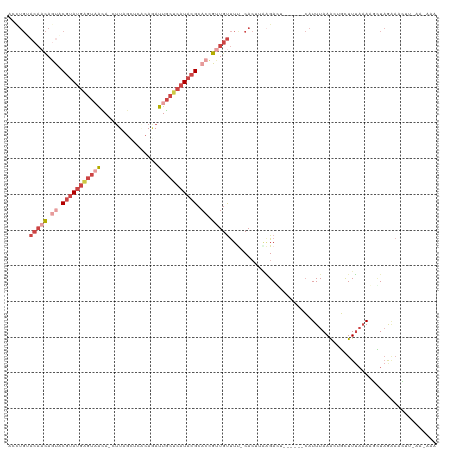
| Location | 11,180,368 – 11,180,481 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.68 |
| Shannon entropy | 0.66024 |
| G+C content | 0.37747 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.79 |
| SVM RNA-class probability | 0.999901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11180368 113 - 22422827 UUUUUUAAAGUUCCUAGCUUUUCAAGCUGUUUAAAUG------ACUCGCAAGUG-GAAACAACGGUGCGUAUGAGCAAUCUGUGAGCUAAGAUGUUACUCAUACGUACUGUUGUCCAAGC ..........(((((((((.....)))))........------...(....).)-)))(((((((((((((((((..((((........))))....)))))))))))))))))...... ( -36.40, z-score = -3.62, R) >droAna3.scaffold_13248 2736543 102 - 4840945 CUACGCCAGAGGACGACUUUAAAAAGGACUUUAAAAC----AAGGUCCUGGUUG-GAGGCAACAGUGCGUAUGAGUAAUAUAAUACGUA---UACGCAGUGUGGGCGUUA---------- ..(((((.........((((((..((((((((.....----))))))))..)))-)))...(((.((((((((.(((......))).))---)))))).))).)))))..---------- ( -39.60, z-score = -5.52, R) >droEre2.scaffold_4690 14781171 118 + 18748788 UUUCUUUAAGUUUCUAGCCUUUAAAACGGCUUAAAUGUGCAAAUGUCUUCAGUG-AAAACAACGGUGCGUAUGAGCAAUCUGCGUGCUAAA-UAUAACUCAUACGUGGUGGUGUACAGUU .........((((..((((........)))).))))(((((..........((.-...))..((.((((((((((................-.....)))))))))).)).))))).... ( -25.50, z-score = -0.16, R) >droYak2.chrX 10807558 112 + 21770863 UUUUUUAAAGUUUCUAGCCUUUAAAGCUGUUUAAAUG------AGUCUUCAGUG-AAAACAACGGUGCGUAUGAGCAAUCUGAAUGCUAAG-UAUUACUCAUACGUACUGUUGUACAGUU ...(((((((........)))))))(((((.......------..((......)-)..(((((((((((((((((.(((((........))-.))).)))))))))))))))))))))). ( -34.50, z-score = -4.20, R) >droSec1.super_15 1865649 105 - 1954846 ---------UUUUUUAAAGUUUAAAACACUUUAAAUG------ACUCUCAAGUGAAAAACAACGGUGCGUAUGAGCAAUUUGUGAGCUAAGAUGUUACCCAUACGUACUGUUGUACAGGU ---------...((((((((.......))))))))..------...............(((((((((((((((((((.((((.....)))).))))...)))))))))))))))...... ( -26.90, z-score = -2.34, R) >droSim1.chrX_random 3137271 105 - 5698898 ---------UUUUUUAAAGUUUAAAGCACUUUAAAUG------ACUCUGAAGUGAAAAACAACGGUGCGUAUGAGCAAUUUGUGAGCUAAGAUGUUACUCAUACGCACUGUUGUGCAGGU ---------.................(((((((....------....)))))))....(((((((((((((((((..((((........))))....)))))))))))))))))...... ( -34.70, z-score = -3.92, R) >consensus UUU_UU_AAGUUUCUAGCCUUUAAAGCACUUUAAAUG______ACUCUUAAGUG_AAAACAACGGUGCGUAUGAGCAAUCUGUGAGCUAAG_UAUUACUCAUACGUACUGUUGUACAGGU ..........................................................(((((((((((((((((.(((..............))).)))))))))))))))))...... (-17.55 = -18.52 + 0.98)

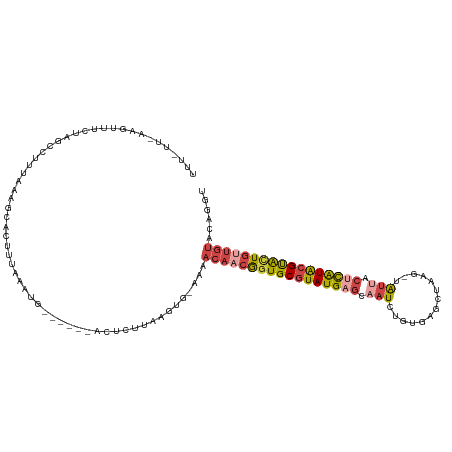
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:48 2011