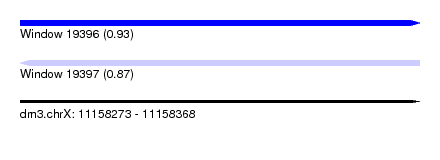
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,158,273 – 11,158,368 |
| Length | 95 |
| Max. P | 0.934587 |

| Location | 11,158,273 – 11,158,368 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 63.29 |
| Shannon entropy | 0.61757 |
| G+C content | 0.42777 |
| Mean single sequence MFE | -21.71 |
| Consensus MFE | -12.65 |
| Energy contribution | -13.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
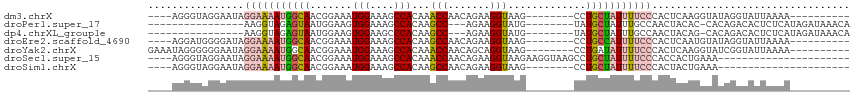
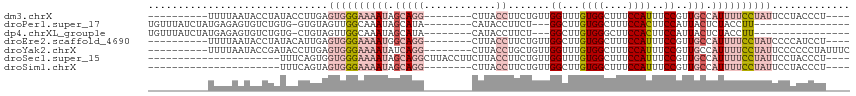
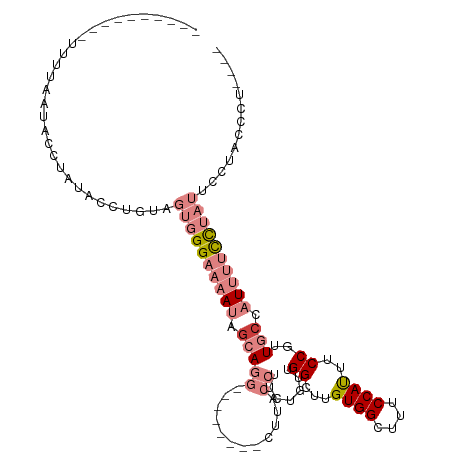
>dm3.chrX 11158273 95 + 22422827 ----AGGGUAGGAAUAGGAAAAUGGCAACGGAAAUGGAAAGCCACAAACCAACAGAAGGUAAG--------CCUGCUAUUUUCCCACUCAAGGUAUAGGUAUUAAAA---------- ----.((((.((.....(((((((((...((...(((....)))....))......(((....--------))))))))))))))))))..................---------- ( -20.60, z-score = -0.49, R) >droPer1.super_17 1723652 89 - 1930428 ----------------AAGGUAGAGUAAUGGAAGUGGAAAGCCACAAGCC---AGAAGGUAUG--------UAUGCUAUUUGCCAACUACAC-CACAGACACUCUCAUAGAUAAACA ----------------..(((..(((..(((..((((....))))...))---)...((((.(--------((...))).)))).)))..))-)....................... ( -17.50, z-score = -0.32, R) >dp4.chrXL_group1e 9249266 89 - 12523060 ----------------AAGGUAGAGUAAUGGAAGUGGAAGCCCACAAGCC---AGAAGGUAUG--------UAUGCUAUUUGCCAACUACAG-CACAGACACUCUCAUAGAUAAACA ----------------...........(((..((((..(((..(((.(((---....))).))--------)..)))...(((........)-))....))))..)))......... ( -17.70, z-score = -0.02, R) >droEre2.scaffold_4690 14760589 95 - 18748788 ----AGGAUGGGGAUAGGAAAAUGGCAACGGAAAUGGAAAGCCACAAGCCAACAGAAGGUAAG--------CCUGCCAUUUUCCCACUCAAUGUAUAGGUAUUAAAA---------- ----....((((....((((((((((...((...(((....)))....))......(((....--------)))))))))))))..)))).................---------- ( -26.40, z-score = -1.79, R) >droYak2.chrX 10789159 99 - 21770863 GAAAUAGGGGGGAAUAGGAAAAUGGCAACGGAAAUGGAAAGCCACAAACCAACAGCAGGUAAG--------CCUGAUAUUUUCCCACUCAAGGUAUCGGUAUUAAAA---------- .......(((..((((((....((((..((....))....))))....)).....((((....--------)))).))))..)))......................---------- ( -22.30, z-score = -0.93, R) >droSec1.super_15 1847241 91 + 1954846 ----AGGGUAGGAAUAGGAAAAUGGCAACGGAAAUGGAAAGCCACAAACCAACAGAAGGUAAGAAGGUAAGCCUGCUAUUUUCCCACCACUGAAA---------------------- ----..(((.((....((((((((((...((...(((....)))....))..............(((....)))))))))))))..)))))....---------------------- ( -22.10, z-score = -1.16, R) >droSim1.chrX 8620230 83 + 17042790 ----AGGGUAGGAAUAGGAAAAUGGCAACGGAAAUGGAAAGCCACAAGCCAACAGAAGGUAAG--------CCUGCUAUUUUCCCACUACUGAAA---------------------- ----..(((((.....((((((((((...((...(((....)))....))......(((....--------)))))))))))))..)))))....---------------------- ( -25.40, z-score = -2.70, R) >consensus ____AGGGUAGGAAUAGGAAAAUGGCAACGGAAAUGGAAAGCCACAAGCCAACAGAAGGUAAG________CCUGCUAUUUUCCCACUACAGGUAUAGGUAUUAAAA__________ ................(((((((((((.......(((....)))...(((.......))).............)))))))))))................................. (-12.65 = -13.24 + 0.60)
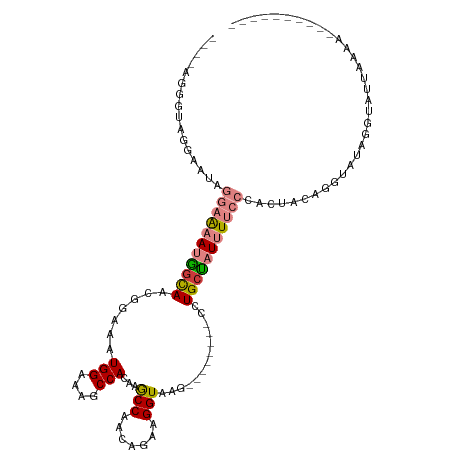
| Location | 11,158,273 – 11,158,368 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.29 |
| Shannon entropy | 0.61757 |
| G+C content | 0.42777 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -8.52 |
| Energy contribution | -10.34 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11158273 95 - 22422827 ----------UUUUAAUACCUAUACCUUGAGUGGGAAAAUAGCAGG--------CUUACCUUCUGUUGGUUUGUGGCUUUCCAUUUCCGUUGCCAUUUUCCUAUUCCUACCCU---- ----------..................((((((((((((.(((((--------((.((.....)).)))))))(((..............))))))))))))))).......---- ( -23.74, z-score = -2.45, R) >droPer1.super_17 1723652 89 + 1930428 UGUUUAUCUAUGAGAGUGUCUGUG-GUGUAGUUGGCAAAUAGCAUA--------CAUACCUUCU---GGCUUGUGGCUUUCCACUUCCAUUACUCUACCUU---------------- ............((((((...(((-(...(((.((.....(((.((--------((..((....---))..)))))))..))))).)))))))))).....---------------- ( -21.20, z-score = -0.81, R) >dp4.chrXL_group1e 9249266 89 + 12523060 UGUUUAUCUAUGAGAGUGUCUGUG-CUGUAGUUGGCAAAUAGCAUA--------CAUACCUUCU---GGCUUGUGGGCUUCCACUUCCAUUACUCUACCUU---------------- ...........(((.((((.((((-((((.........))))))))--------.))))....(---((...((((....))))..)))...)))......---------------- ( -20.30, z-score = -0.26, R) >droEre2.scaffold_4690 14760589 95 + 18748788 ----------UUUUAAUACCUAUACAUUGAGUGGGAAAAUGGCAGG--------CUUACCUUCUGUUGGCUUGUGGCUUUCCAUUUCCGUUGCCAUUUUCCUAUCCCCAUCCU---- ----------..................(..(((((((((((((((--------((.((.....)).)))))((((....)))).......))))))))))))..).......---- ( -25.80, z-score = -2.29, R) >droYak2.chrX 10789159 99 + 21770863 ----------UUUUAAUACCGAUACCUUGAGUGGGAAAAUAUCAGG--------CUUACCUGCUGUUGGUUUGUGGCUUUCCAUUUCCGUUGCCAUUUUCCUAUUCCCCCCUAUUUC ----------..................((((((((((((...(((--------....)))((...(((...((((....))))..)))..)).))))))))))))........... ( -22.50, z-score = -1.91, R) >droSec1.super_15 1847241 91 - 1954846 ----------------------UUUCAGUGGUGGGAAAAUAGCAGGCUUACCUUCUUACCUUCUGUUGGUUUGUGGCUUUCCAUUUCCGUUGCCAUUUUCCUAUUCCUACCCU---- ----------------------....((.(((((((((((((.(((............))).)))))((...(((((..............)))))...))..))))))))))---- ( -25.74, z-score = -2.84, R) >droSim1.chrX 8620230 83 - 17042790 ----------------------UUUCAGUAGUGGGAAAAUAGCAGG--------CUUACCUUCUGUUGGCUUGUGGCUUUCCAUUUCCGUUGCCAUUUUCCUAUUCCUACCCU---- ----------------------....((.(((((((((((.(((((--------((.((.....)).)))))))(((..............)))))))))))))).)).....---- ( -24.54, z-score = -2.83, R) >consensus __________UUUUAAUACCUAUACCUGUAGUGGGAAAAUAGCAGG________CUUACCUUCUGUUGGCUUGUGGCUUUCCAUUUCCGUUGCCAUUUUCCUAUUCCUACCCU____ ..............................((((((((((.(((..............((.......))...((((....))))......))).))))))))))............. ( -8.52 = -10.34 + 1.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:41 2011