| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,154,940 – 11,155,036 |
| Length | 96 |
| Max. P | 0.801335 |

| Location | 11,154,940 – 11,155,036 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 64.26 |
| Shannon entropy | 0.61172 |
| G+C content | 0.49485 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -7.48 |
| Energy contribution | -7.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
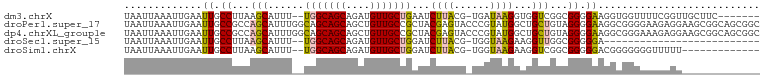
>dm3.chrX 11154940 96 + 22422827 -------GAAGCAACCGAAAACCACCUUCCCCGCCGACCACCUUAUCA-CGUAAGAUUCAGCAACAUCUGCUGCCA--AAAUGCUUAAGGCAAUUCAAUUUAAUUA -------((((..............))))...(((......(((((..-.)))))...(((((.....)))))...--..........)))............... ( -12.04, z-score = -0.60, R) >droPer1.super_17 1720533 106 - 1930428 GCCGCUGCCGCUUCCUCUUCCCCGCCUUCCCCUACAGCAGCCAUACGGGUACUCGUAGCGGCAACAGCUGCUGCUGCCAAAUGCUGGCGGCAAUUCAAUUUAAUUA (((((....((............)).........((((((((.....)))....(((((((((.....)))))))))....))))))))))............... ( -33.60, z-score = -1.15, R) >dp4.chrXL_group1e 9246176 106 - 12523060 GCCGCUGCCGCUUCCUCUUUCCCGCCUUCCCCUACAGCAGCCAUACGGGUACUCGUAGCGGCAACAGCUGCUGCUGCCAAAUGCUGGCGGCAAUUCAAUUUAAUUA ((((((((((((........((((.....................)))).......))))))).((((.((....)).....)))))))))............... ( -33.86, z-score = -1.26, R) >droSec1.super_15 1844035 77 + 1954846 --------------------------UCCCCCGCCAACCUUCUUACCA-CGUAAGAUCCAGCAACAUCUGCUGCCA--AAAUGCUUAAGGCAAUUCAAUUUAAUUA --------------------------......(((.....((((((..-.))))))..(((((.....)))))...--..........)))............... ( -14.80, z-score = -3.52, R) >droSim1.chrX 8617004 90 + 17042790 -------------AAAAACCCCCCCGUCCCCCGCCGACCUUCUUACCA-CGUAAGAUCCAGCAACAUCUGCUGCCA--AAAUGCUUAAGGCAAUUCAAUUUAAUUA -------------...................(((.....((((((..-.))))))..(((((.....)))))...--..........)))............... ( -14.80, z-score = -3.10, R) >consensus _____________ACUCAUCCCCGCCUUCCCCGCCAACCGCCUUACCA_CGUAAGAUCCAGCAACAUCUGCUGCCA__AAAUGCUUAAGGCAAUUCAAUUUAAUUA ..........................................................(((((.....)))))........(((.....))).............. ( -7.48 = -7.24 + -0.24)


| Location | 11,154,940 – 11,155,036 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 64.26 |
| Shannon entropy | 0.61172 |
| G+C content | 0.49485 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -11.76 |
| Energy contribution | -11.56 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11154940 96 - 22422827 UAAUUAAAUUGAAUUGCCUUAAGCAUUU--UGGCAGCAGAUGUUGCUGAAUCUUACG-UGAUAAGGUGGUCGGCGGGGAAGGUGGUUUUCGGUUGCUUC------- ......((((((((..((((..((...(--..(((((....)))))..)((((((..-...)))))).....))....))))..))..)))))).....------- ( -24.90, z-score = -0.95, R) >droPer1.super_17 1720533 106 + 1930428 UAAUUAAAUUGAAUUGCCGCCAGCAUUUGGCAGCAGCAGCUGUUGCCGCUACGAGUACCCGUAUGGCUGCUGUAGGGGAAGGCGGGGAAGAGGAAGCGGCAGCGGC .............((((.(((((...))))).))))..((((((((((((.(.....(((((....((.(....).))...))))).....)..)))))))))))) ( -43.50, z-score = -1.96, R) >dp4.chrXL_group1e 9246176 106 + 12523060 UAAUUAAAUUGAAUUGCCGCCAGCAUUUGGCAGCAGCAGCUGUUGCCGCUACGAGUACCCGUAUGGCUGCUGUAGGGGAAGGCGGGAAAGAGGAAGCGGCAGCGGC .............((((.(((((...))))).))))..((((((((((((.(.....(((((....((.(....).))...))))).....)..)))))))))))) ( -42.40, z-score = -1.78, R) >droSec1.super_15 1844035 77 - 1954846 UAAUUAAAUUGAAUUGCCUUAAGCAUUU--UGGCAGCAGAUGUUGCUGGAUCUUACG-UGGUAAGAAGGUUGGCGGGGGA-------------------------- .............(((((........((--..(((((....)))))..))((((((.-..)))))).....)))))....-------------------------- ( -19.50, z-score = -1.25, R) >droSim1.chrX 8617004 90 - 17042790 UAAUUAAAUUGAAUUGCCUUAAGCAUUU--UGGCAGCAGAUGUUGCUGGAUCUUACG-UGGUAAGAAGGUCGGCGGGGGACGGGGGGGUUUUU------------- ..........(((((.((((..((..((--..(((((....)))))..))((((((.-..))))))......)).(....))))).)))))..------------- ( -24.30, z-score = -1.81, R) >consensus UAAUUAAAUUGAAUUGCCUUAAGCAUUU__UGGCAGCAGAUGUUGCUGCAUCUUACG_UGGUAAGGAGGUUGGCGGGGAAGGCGGGGAAGAGG_____________ .............((.((...(((......(((((((....)))))))...(((((....)))))...)))...)).))........................... (-11.76 = -11.56 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:39 2011