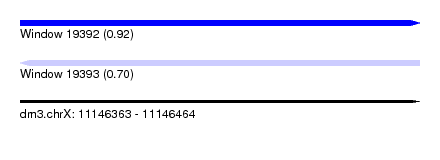
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,146,363 – 11,146,464 |
| Length | 101 |
| Max. P | 0.920976 |

| Location | 11,146,363 – 11,146,464 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.64 |
| Shannon entropy | 0.16032 |
| G+C content | 0.53519 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -28.30 |
| Energy contribution | -29.60 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
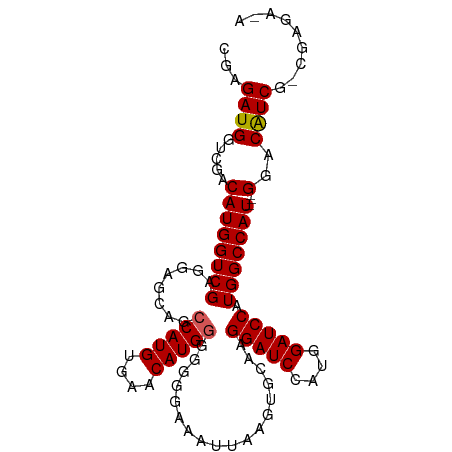
>dm3.chrX 11146363 101 + 22422827 CGAGAUGGUCGACAUGGUCGAGGAGCAGCCAUGUGAACAUGAGGGGGAAAUUAAGUGCAAGGAUCCAUGGAUCCAUGGCCAUCAUGGACAUCAUCGAGAAA (((((((.((.(((((((.(.....).)))))))...(((((...((..........((.(((((....))))).)).)).))))))))))).)))..... ( -30.80, z-score = -1.11, R) >droEre2.scaffold_4690 14749606 89 - 18748788 CGAGAUGGUCGACAUGGUCGAGGAGCAGCCAUGUGAACAUGGGGGG-AAAUUAAGUGCAAGGAUCCAUGGAUCCAUGGCCAU---GGACGUCU-------- ..(((((.....((((((((........(((((....)))))....-.............(((((....))))).)))))))---)..)))))-------- ( -32.00, z-score = -2.08, R) >droYak2.chrX 10774855 90 - 21770863 CGAGAUGGUCGACAUGGUCGAGGAGCAGCCAUGUGAACAUGGAGGGGAAAUUAAGUGCAAGGAUCCAUGGAUCCAUGGCCAU---GGACGUCG-------- ...((((.....((((((((.....(..(((((....)))))..)...............(((((....))))).)))))))---)..)))).-------- ( -32.60, z-score = -2.56, R) >droSec1.super_15 1835648 97 + 1954846 CGAGAUGGUCGACAUGGUCGAGGAGCAGCCAUGUGAACAUGGGGGG-AAAUUAAGUGCAAGGAUCCAUGGAUCCAUGGCCAU---GGACAUCGGCGAGAGA ((.((((.....((((((((........(((((....)))))....-.............(((((....))))).)))))))---)..))))..))..... ( -33.50, z-score = -2.16, R) >droSim1.chrX_random 3133187 98 + 5698898 CGAGAUGGUCGACAUGGUCGAGGAGCAGCCAUGUGAACAUGGGGGGGAAAUUAAGUGCAAGGAUCCAUGGAUCCAUGGCCAU---GGACAUCGACGAGAGA .......(((((((((((((........(((((....)))))..................(((((....))))).)))))))---)....)))))...... ( -33.30, z-score = -2.24, R) >consensus CGAGAUGGUCGACAUGGUCGAGGAGCAGCCAUGUGAACAUGGGGGGGAAAUUAAGUGCAAGGAUCCAUGGAUCCAUGGCCAU___GGACAUCG_CGAGA_A ...((((..(...(((((((....(((.(((((....)))))((......))...)))..(((((....))))).)))))))...)..))))......... (-28.30 = -29.60 + 1.30)

| Location | 11,146,363 – 11,146,464 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Shannon entropy | 0.16032 |
| G+C content | 0.53519 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -21.81 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
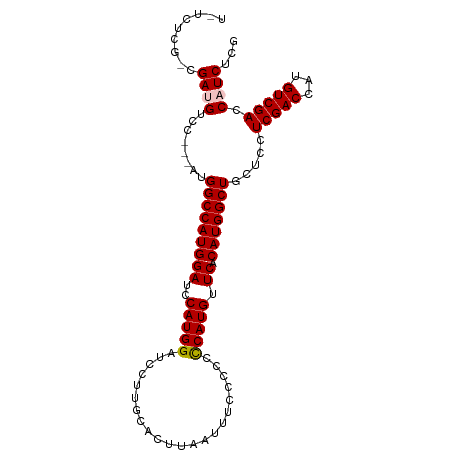
>dm3.chrX 11146363 101 - 22422827 UUUCUCGAUGAUGUCCAUGAUGGCCAUGGAUCCAUGGAUCCUUGCACUUAAUUUCCCCCUCAUGUUCACAUGGCUGCUCCUCGACCAUGUCGACCAUCUCG .....(((.(((((((((((.((.((.(((((....))))).))............)).)))))...(((((((........).)))))).)).))))))) ( -24.72, z-score = -0.81, R) >droEre2.scaffold_4690 14749606 89 + 18748788 --------AGACGUCC---AUGGCCAUGGAUCCAUGGAUCCUUGCACUUAAUUU-CCCCCCAUGUUCACAUGGCUGCUCCUCGACCAUGUCGACCAUCUCG --------....((((---((((.(..(((((....))))).............-....(((((....))))).........).)))))..)))....... ( -21.80, z-score = -0.97, R) >droYak2.chrX 10774855 90 + 21770863 --------CGACGUCC---AUGGCCAUGGAUCCAUGGAUCCUUGCACUUAAUUUCCCCUCCAUGUUCACAUGGCUGCUCCUCGACCAUGUCGACCAUCUCG --------(((.....---..(((((((((..((((((....................)))))).)).))))))).....(((((...))))).....))) ( -22.05, z-score = -1.12, R) >droSec1.super_15 1835648 97 - 1954846 UCUCUCGCCGAUGUCC---AUGGCCAUGGAUCCAUGGAUCCUUGCACUUAAUUU-CCCCCCAUGUUCACAUGGCUGCUCCUCGACCAUGUCGACCAUCUCG .........(((((((---((((.(..(((((....))))).............-....(((((....))))).........).)))))..)).))))... ( -24.00, z-score = -1.33, R) >droSim1.chrX_random 3133187 98 - 5698898 UCUCUCGUCGAUGUCC---AUGGCCAUGGAUCCAUGGAUCCUUGCACUUAAUUUCCCCCCCAUGUUCACAUGGCUGCUCCUCGACCAUGUCGACCAUCUCG ......(((((....(---((((.(..(((((....)))))..................(((((....))))).........).))))))))))....... ( -25.30, z-score = -1.76, R) >consensus U_UCUCG_CGAUGUCC___AUGGCCAUGGAUCCAUGGAUCCUUGCACUUAAUUUCCCCCCCAUGUUCACAUGGCUGCUCCUCGACCAUGUCGACCAUCUCG .........((((........(((((((((..(((((......................))))).)).))))))).....(((((...))))).))))... (-21.81 = -22.05 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:38 2011