
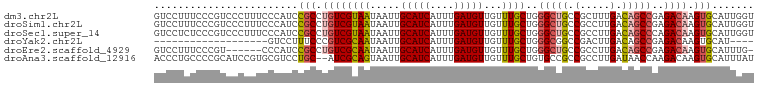
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 968,555 – 968,655 |
| Length | 100 |
| Max. P | 0.826377 |

| Location | 968,555 – 968,655 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Shannon entropy | 0.40798 |
| G+C content | 0.51726 |
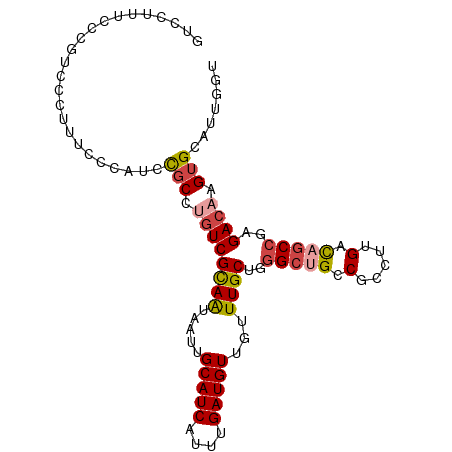
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -17.13 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 968555 100 + 23011544 GUCCUUUCCCGUCCCUUUCCCAUCCGCCUGUCGUAAUAAUUGCAUCAUUUGAUGUUGUUUGCUGGGCUGCCGCUUUGACAGCCGAGACAAGUGCAUUGGU ...................(((..(((.((((((((.....(((((....)))))...))))..(((((.(.....).)))))..)))).)))...))). ( -25.40, z-score = -1.18, R) >droSim1.chr2L 959992 100 + 22036055 GUCCUUUCCCGUCCCUUUCCCAUCCGCCUGUCGUAAUAAUUGCAUCAUUUGAUGUUGUUUGCUGGGCUGCCGCCUUGACAGCCGAGACAAGUGCAUUGGU ...................(((..(((.((((((((.....(((((....)))))...))))..(((((.(.....).)))))..)))).)))...))). ( -25.40, z-score = -1.20, R) >droSec1.super_14 939529 100 + 2068291 GUCCUCUCCCGUCCCUUUCCCAUCCGCCUGUCGUAAUAAUUGCAUCAUUUGAUGUUGUUUGCUGGGCUGCCGCCUUGACAGCCCAGACAAGUGCAUUGGU ...................(((..(((.((((((((.....(((((....)))))...))))(((((((.(.....).))))))))))).)))...))). ( -29.50, z-score = -2.87, R) >droYak2.chr2L 956937 77 + 22324452 -------------------GUCCUUUCCCGUCGCAAUAAUUGCAUCAUUUGAUGUUGUUUGCUGGGCGGCCGACUUGACAGCCGAGACAAGUGCAU---- -------------------(((....((((..((((.....(((((....)))))...))))))))((((.(......).)))).)))........---- ( -21.00, z-score = -0.41, R) >droEre2.scaffold_4929 1028248 93 + 26641161 GUCCUUUCCCGU------CCCAUCCGCCUGUCGCAAUAAUUGCAUCAUUUGAUGUUGUUUGCUGGGCUGCCGCCUUGACAGCCGAGACAAGUGCAUUUG- (..(((....((------(.....((.(((((((((.....(((((....)))))...)))).((((....)))).))))).)).))))))..).....- ( -24.30, z-score = -1.19, R) >droAna3.scaffold_12916 2687994 98 + 16180835 ACCCUGCCCCGCAUCCGUGCGUCCUGC--AUCGCAGUAAUUGCAUCAUUUGAUGUUGUUUGCUGUGCCGCCGCCUUGAUAACCAAGACAAGUGCAUUUAU ..........((((..(((((....((--..((((((((..(((((....)))))...))))))))..)))))((((.....))))))..))))...... ( -27.00, z-score = -2.12, R) >consensus GUCCUUUCCCGUCCCUUUCCCAUCCGCCUGUCGCAAUAAUUGCAUCAUUUGAUGUUGUUUGCUGGGCUGCCGCCUUGACAGCCGAGACAAGUGCAUUGGU ........................(((.((((((((.....(((((....)))))...))))..(((((.(.....).)))))..)))).)))....... (-17.13 = -17.72 + 0.59)
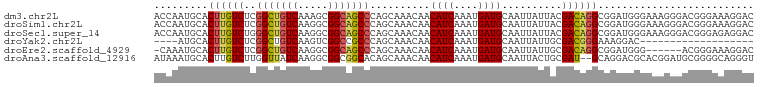
| Location | 968,555 – 968,655 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Shannon entropy | 0.40798 |
| G+C content | 0.51726 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 968555 100 - 23011544 ACCAAUGCACUUGUCUCGGCUGUCAAAGCGGCAGCCCAGCAAACAACAUCAAAUGAUGCAAUUAUUACGACAGGCGGAUGGGAAAGGGACGGGAAAGGAC .((.((.(.((((((..(((((((.....)))))))..........((((....))))..........)))))).).)).)).................. ( -27.20, z-score = -1.32, R) >droSim1.chr2L 959992 100 - 22036055 ACCAAUGCACUUGUCUCGGCUGUCAAGGCGGCAGCCCAGCAAACAACAUCAAAUGAUGCAAUUAUUACGACAGGCGGAUGGGAAAGGGACGGGAAAGGAC .((.((.(.((((((..(((((((.....)))))))..........((((....))))..........)))))).).)).)).................. ( -27.20, z-score = -1.13, R) >droSec1.super_14 939529 100 - 2068291 ACCAAUGCACUUGUCUGGGCUGUCAAGGCGGCAGCCCAGCAAACAACAUCAAAUGAUGCAAUUAUUACGACAGGCGGAUGGGAAAGGGACGGGAGAGGAC .((.((.(.(((((((((((((((.....)))))))))........((((....))))..........)))))).).)).)).................. ( -31.90, z-score = -2.55, R) >droYak2.chr2L 956937 77 - 22324452 ----AUGCACUUGUCUCGGCUGUCAAGUCGGCCGCCCAGCAAACAACAUCAAAUGAUGCAAUUAUUGCGACGGGAAAGGAC------------------- ----.....((((((.((((((......))))))....((((....((((....))))......)))))))))).......------------------- ( -22.20, z-score = -1.22, R) >droEre2.scaffold_4929 1028248 93 - 26641161 -CAAAUGCACUUGUCUCGGCUGUCAAGGCGGCAGCCCAGCAAACAACAUCAAAUGAUGCAAUUAUUGCGACAGGCGGAUGGG------ACGGGAAAGGAC -........(((((((((.(((((..(((....)))..((((....((((....))))......))))....))))).))))------)))))....... ( -29.10, z-score = -1.95, R) >droAna3.scaffold_12916 2687994 98 - 16180835 AUAAAUGCACUUGUCUUGGUUAUCAAGGCGGCGGCACAGCAAACAACAUCAAAUGAUGCAAUUACUGCGAU--GCAGGACGCACGGAUGCGGGGCAGGGU .........(((((((((..((((...(((...(((..(((.....((((....)))).......)))..)--))....)))...))))))))))))).. ( -28.90, z-score = -1.42, R) >consensus ACCAAUGCACUUGUCUCGGCUGUCAAGGCGGCAGCCCAGCAAACAACAUCAAAUGAUGCAAUUAUUACGACAGGCGGAUGGGAAAGGGACGGGAAAGGAC .........((((((..(((((((.....)))))))..........((((....))))..........)))))).......................... (-17.76 = -18.10 + 0.34)
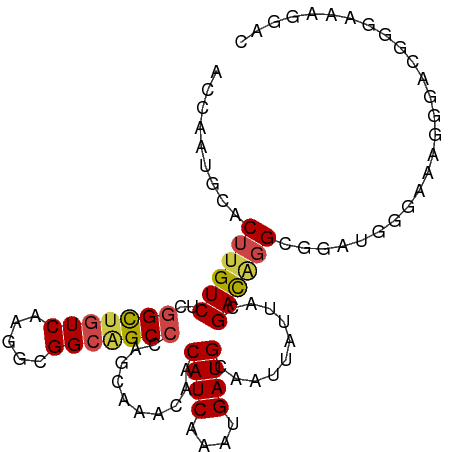
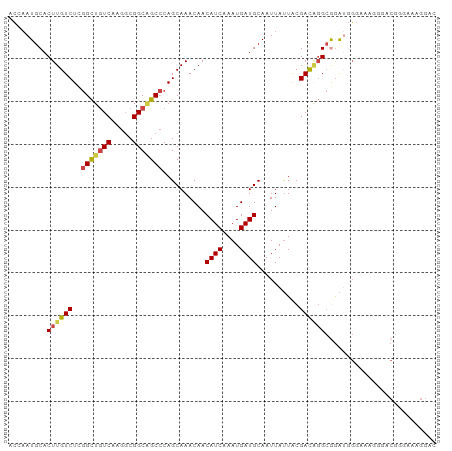
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:26 2011