| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,139,927 – 11,140,020 |
| Length | 93 |
| Max. P | 0.999553 |

| Location | 11,139,927 – 11,140,020 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 64.29 |
| Shannon entropy | 0.62653 |
| G+C content | 0.34584 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -4.95 |
| Energy contribution | -5.48 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
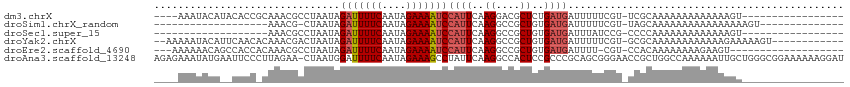
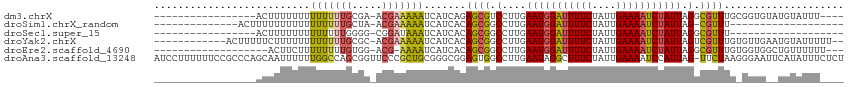
>dm3.chrX 11139927 93 + 22422827 ----AAAUACAUACACCGCAAACGCCUAAUAGAUUUUCAAUAGAAAAUCCAUUCAAGGACGCUCUGAUGAUUUUUCGU-UCGCAAAAAAAAAAAAAGU----------------- ----.............((...((((((((.(((((((....))))))).)))..))).))....(((((....))))-).))...............----------------- ( -14.50, z-score = -1.83, R) >droSim1.chrX_random 3127203 80 + 5698898 -------------------AAACG-CUAAUAGAUUUUCAAUAGAAAAUCCAUUCAAGGCCGCUGUGAUGAUUUUUCGU-UAGCAAAAAAAAAAAAAAAAGU-------------- -------------------....(-(((((.(((((((....)))))))((((..((....))..)))).......))-))))..................-------------- ( -13.90, z-score = -1.76, R) >droSec1.super_15 1829724 78 + 1954846 -------------------AAACGCCUAAUAGAUUUUCAAUAGAAAAUCCAUUCAAGGCCGCUGUGAUGAUUUAUCCG-CCCCAAAAAAAAAAAAAGU----------------- -------------------....(((((((.(((((((....))))))).)))..)))).((.(.((((...))))))-)..................----------------- ( -12.40, z-score = -2.34, R) >droYak2.chrX 10769148 100 - 21770863 --AAAAAUACAUUCAACACAAACGACUAAUAGAUUUUCAAUAGAAAAUCCAUUCAAGGCCGCUGUGAUGAUUUUUCGU-GCGCAAAAAAAAAAAAGAAAAAGU------------ --..........(((.((((..((.(((((.(((((((....))))))).)))..))..)).)))).)))((((((..-................))))))..------------ ( -14.17, z-score = -1.02, R) >droEre2.scaffold_4690 14743360 91 - 18748788 ---AAAAAACAGCCACCACAAACGCCUAAUAGAUUUUCAAUAGAAAAUCCAUUCAAGGCCGCUGUGAUGAUUUU-CGU-CCACAAAAAAAAGAAGU------------------- ---.....(((((..........(((((((.(((((((....))))))).)))..)))).)))))((((.....-)))-)................------------------- ( -17.60, z-score = -3.27, R) >droAna3.scaffold_13248 2699289 114 + 4840945 AGAGAAAUAUGAAUUCCCUUAGAA-CUAAUGGAUUUUCAAUAGAAAGCCUAUUCAAGGCCACUCCGCCCGCAGCGGGAACCGCUGGCCAAAAAAUUGCUGGGCGGAAAAAAGGAU .........((((.(((.(((...-.))).)))..)))).......((((.....))))...(((((((.((((((...))))))((.........)).)))))))......... ( -34.10, z-score = -1.66, R) >consensus ____AAAUACA__CACC_CAAACGCCUAAUAGAUUUUCAAUAGAAAAUCCAUUCAAGGCCGCUGUGAUGAUUUUUCGU_CCGCAAAAAAAAAAAAAGA_________________ ...............................(((((((....)))))))((((..((....))..)))).............................................. ( -4.95 = -5.48 + 0.53)
| Location | 11,139,927 – 11,140,020 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.29 |
| Shannon entropy | 0.62653 |
| G+C content | 0.34584 |
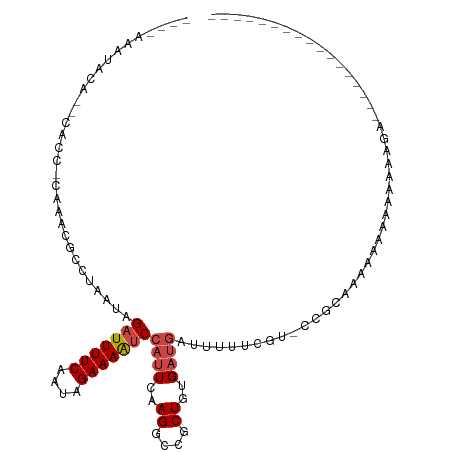
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -12.21 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11139927 93 - 22422827 -----------------ACUUUUUUUUUUUUUGCGA-ACGAAAAAUCAUCAGAGCGUCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUGCGGUGUAUGUAUUU---- -----------------.........(((((((...-.))))))).((((.(((((.(((..(((((((((((....)))))))))))))))))))..)))).........---- ( -24.50, z-score = -3.01, R) >droSim1.chrX_random 3127203 80 - 5698898 --------------ACUUUUUUUUUUUUUUUUGCUA-ACGAAAAAUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAG-CGUUU------------------- --------------............(((((((...-.)))))))...........((....(((((((((((....))))))))))).)-)....------------------- ( -14.40, z-score = -1.68, R) >droSec1.super_15 1829724 78 - 1954846 -----------------ACUUUUUUUUUUUUUGGGG-CGGAUAAAUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUU------------------- -----------------..................(-(.(((.....)))...)).((((..(((((((((((....)))))))))))))))....------------------- ( -19.60, z-score = -2.84, R) >droYak2.chrX 10769148 100 + 21770863 ------------ACUUUUUCUUUUUUUUUUUUGCGC-ACGAAAAAUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGUCGUUUGUGUUGAAUGUAUUUUU-- ------------..............(((((((...-.)))))))(((.(((((((((....(((((((((((....))))))))))).)))).))))).)))..........-- ( -25.50, z-score = -3.83, R) >droEre2.scaffold_4690 14743360 91 + 18748788 -------------------ACUUCUUUUUUUUGUGG-ACG-AAAAUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUGUGGUGGCUGUUUUUU--- -------------------........((((((...-.))-))))(((((((((..((((..(((((((((((....)))))))))))))))..))))))))).........--- ( -28.80, z-score = -4.41, R) >droAna3.scaffold_13248 2699289 114 - 4840945 AUCCUUUUUUCCGCCCAGCAAUUUUUUGGCCAGCGGUUCCCGCUGCGGGCGGAGUGGCCUUGAAUAGGCUUUCUAUUGAAAAUCCAUUAG-UUCUAAGGGAAUUCAUAUUUCUCU .((((((.((((((((..(((....)))((.(((((...))))))))))))))).(((((.....)))))..(((.((......)).)))-....)))))).............. ( -37.00, z-score = -1.96, R) >consensus _________________ACUUUUUUUUUUUUUGCGG_ACGAAAAAUCAUCACAGCGGCCUUGAAUGGAUUUUCUAUUGAAAAUCUAUUAGGCGUUUG_GGUG__UGUAUUU____ ..........................(((((((.....))))))).......((((.(....(((((((((((....))))))))))).).)))).................... (-12.21 = -12.22 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:36 2011