| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,139,635 – 11,139,738 |
| Length | 103 |
| Max. P | 0.999260 |

| Location | 11,139,635 – 11,139,738 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 56.06 |
| Shannon entropy | 0.78901 |
| G+C content | 0.33687 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -8.35 |
| Energy contribution | -9.99 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
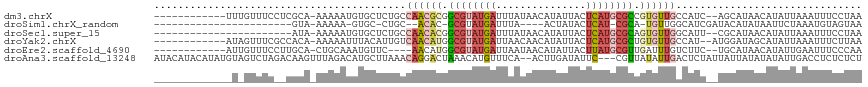
>dm3.chrX 11139635 103 - 22422827 ------------UUUGUUUCCUCGCA-AAAAAUGUGCUCUGCCAACGCGGCGUAUGAUUUAUAACAUAUUACUCAUGCGCCGUGUUGCCAUC--AGCAUAACAUAUUAAAUUUCCUAA ------------(((((......)))-))...((((((.((.(((((((((((((((...(((...)))...))))))))))))))).))..--)))))).................. ( -33.50, z-score = -5.77, R) >droSim1.chrX_random 3126979 83 - 5698898 -----------------------GUA-AAAAA-GUGC-CUGC--ACAC-GCGUAUGAUUUA----ACUAUACUCAU-CGCA-UGUUGGCAUCGAUACAUAUAAUUCUAAAUGUAGUAA -----------------------...-.....-((((-(...--(((.-(((.((((....----.......))))-))).-))).)))))...(((((.((....)).))))).... ( -16.10, z-score = -0.81, R) >droSec1.super_15 1829473 92 - 1954846 -----------------------AUA-AAAAAUGUGCUCUGCCAACACGGCGUAUGAUUUAUAACAUAUUACUCAUGCGCAGUGUUGGCAUU--CGCAUAACAUAUUAAAUUUCCUAA -----------------------...-.....(((((..(((((((((.((((((((...(((...)))...)))))))).)))))))))..--.))))).................. ( -31.80, z-score = -5.84, R) >droYak2.chrX 10768857 103 + 21770863 ------------AUAGUUUCGCCACA-AAAAAUUUACAUUGUCAACAUGGCGUAUGAUUAACAACAUAUUACUCAUGCGCUGUGUUGCCAU--AUGGAUAGCAUAUUAAAUUUCUUAA ------------..............-..(((((((...((((((((((((((((((.(((.......))).)))))))))))))))((..--..))...)))...)))))))..... ( -24.50, z-score = -2.45, R) >droEre2.scaffold_4690 14743064 99 + 18748788 ------------AUUGUUUCCUUGCA-CUGCAAAUGUUC----AACAUGGCGUAUGAUUAAUAACAUAUUACUUAUGCGUUGAUUUGUCUUC--UGCAUAACAUAUUGAAUUUCCCAA ------------..((((....((((-..(((((((...----..)..(((((((((.(((((...))))).))))))))).))))))....--)))).))))............... ( -16.30, z-score = -0.44, R) >droAna3.scaffold_13248 2697950 113 - 4840945 AUACAUACAUAUGUAGUCUAGACAAGUUUAGACAUGCUUAAACAGGACUAAACAUGUUUCA--ACUUGAUAUUC---CGUUAUAUUGACUCUAUUAUUAUAUAUAUUGACCUCUCUCU ...((...(((((((((.((((((((((.(((((((.(((........))).))))))).)--)))))......---.(((.....)))))))..)))))))))..)).......... ( -20.90, z-score = -1.67, R) >consensus _____________U_GUUUC__CACA_AAAAAUGUGCUCUGCCAACACGGCGUAUGAUUUAUAACAUAUUACUCAUGCGCUGUGUUGCCAUC__UGCAUAACAUAUUAAAUUUCCUAA ..........................................((((((.((((((((...............)))))))).))))))............................... ( -8.35 = -9.99 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:35 2011