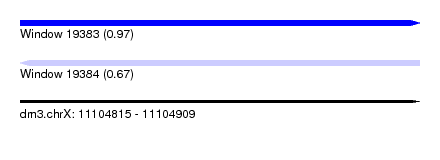
| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,104,815 – 11,104,909 |
| Length | 94 |
| Max. P | 0.972125 |

| Location | 11,104,815 – 11,104,909 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 61.86 |
| Shannon entropy | 0.71837 |
| G+C content | 0.36003 |
| Mean single sequence MFE | -11.26 |
| Consensus MFE | -5.88 |
| Energy contribution | -5.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
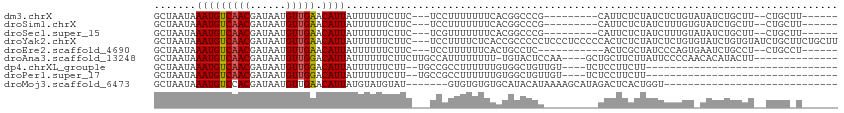
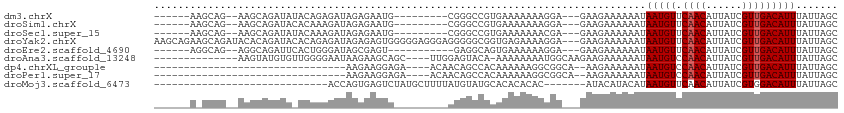
>dm3.chrX 11104815 94 + 22422827 GCUAAUAAAUGUCAACGAUAAUGUUGAACAUUAUUUUUUCUUC---UCCUUUUUUUCACGGCCCG---------CAUUCUCUAUCUCUGUAUAUCUGCUU--CUGCUU------ ((..((((((((((((......)))).)))))...........---...........((((....---------............)))).)))..))..--......------ ( -8.29, z-score = -0.30, R) >droSim1.chrX 8589121 94 + 17042790 GCUAAUAAAUGUCAACGAUAAUGUUGAACAUUAUUUUUUCUUC---UCCUUUUUUUCACGGCCCG---------CAUUCUCUAUCUUUGUGUAUCUGCUU--CUGCUU------ ((.....(((((((((......)))).)))))...........---.............((..((---------((...........))))..)).))..--......------ ( -10.40, z-score = -0.87, R) >droSec1.super_15 1796056 94 + 1954846 GCUAAUAAAUGUCAACGAUAAUGUUGAACAUUAUUUUUUCUUC---UCGUUUUUUUCACGGCCCG---------CAUUCUCUAUCUUUGUAUAUCUGCUU--CUGCUU------ ((.....(((((((((......)))).)))))...........---((((.......))))...)---------).....................((..--..))..------ ( -9.70, z-score = -0.45, R) >droYak2.chrX 10734904 111 - 21770863 GCUAAUAAAUGUCAACGAUAAUGUUGAACAUUAUUUUUUCUUC---UCCUUUUCUCACCGCCCCCUCCCUCCCCCACUCUCUAUCUCUGUGUAUCUGUGUAUCUGCUUCUGCUU ((..((((((((((((......)))).)))))...........---............................(((...........)))........)))..))........ ( -9.10, z-score = -2.13, R) >droEre2.scaffold_4690 14708962 92 - 18748788 GCUAAUAAAUGUCAACGAUAAUGUUGAACAUUAUUUUUUCUUC---UCCUUUUUUCACUGCCUC-----------ACUCGCUAUCCCAGUGAAUCUGCCU--CUGCCU------ ((.....(((((((((......)))).)))))...........---.......(((((((....-----------...........)))))))...))..--......------ ( -12.96, z-score = -2.49, R) >droAna3.scaffold_13248 2653528 95 + 4840945 GCUAAUAAAUGUCAACGAUAAUGUUGGACAUUAUUUUUUCUUCUUGCCAUUUUUUUU-UGUACUCCAA----GCUGCUUCUUAUUCCCCAACACAUACUU-------------- (((....(((((((((......))).))))))...............((........-)).......)----))..........................-------------- ( -7.40, z-score = 0.41, R) >dp4.chrXL_group1e 9191758 76 - 12523060 GCUAAUAAAUGUCAACGAUAAUGUUGGACAUUAUUUUUUCUU--UGCCGCCUUUUUUGUGGCUGUUGU----UCUCCUUCUU-------------------------------- ............(((((((((((.....))))))).......--.(((((.......))))).)))).----..........-------------------------------- ( -13.70, z-score = -2.17, R) >droPer1.super_17 1662072 76 - 1930428 GCUAAUAAAUGUCAACGAUAAUGUUGGACAUUAUUUUUUCUU--UGCCGCCUUUUUUGUGGCUGUUGU----UCUCCUUCUU-------------------------------- ............(((((((((((.....))))))).......--.(((((.......))))).)))).----..........-------------------------------- ( -13.70, z-score = -2.17, R) >droMoj3.scaffold_6473 10195715 78 + 16943266 GCUAAUAAAUGUCCACGAUAAUGUUGAACAUUAUGUAUGUAU-------GUGUGUGUGCAUACAUAAAAGCAUAGACUCACUGGU----------------------------- (((......((((.((......)).).)))((((((((((((-------......)))))))))))).)))..............----------------------------- ( -16.10, z-score = 0.08, R) >consensus GCUAAUAAAUGUCAACGAUAAUGUUGAACAUUAUUUUUUCUUC___UCCUUUUUUUUACGGCCCG_________CACUCUCUAUCUCUGU_UAUCUGCUU__CUGC_U______ .......(((((((((......))))).)))).................................................................................. ( -5.88 = -5.77 + -0.11)
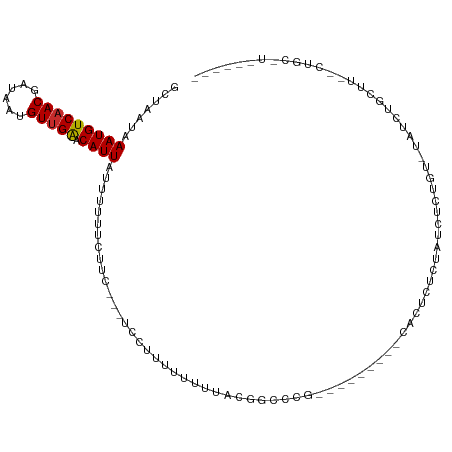
| Location | 11,104,815 – 11,104,909 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 61.86 |
| Shannon entropy | 0.71837 |
| G+C content | 0.36003 |
| Mean single sequence MFE | -12.34 |
| Consensus MFE | -5.56 |
| Energy contribution | -5.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
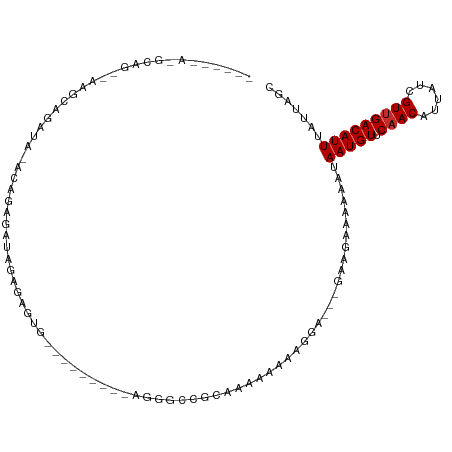
>dm3.chrX 11104815 94 - 22422827 ------AAGCAG--AAGCAGAUAUACAGAGAUAGAGAAUG---------CGGGCCGUGAAAAAAAGGA---GAAGAAAAAAUAAUGUUCAACAUUAUCGUUGACAUUUAUUAGC ------..((..--..(((..(((......))).....))---------)..))..............---........((((((((.((((......))))))).)))))... ( -10.00, z-score = 0.26, R) >droSim1.chrX 8589121 94 - 17042790 ------AAGCAG--AAGCAGAUACACAAAGAUAGAGAAUG---------CGGGCCGUGAAAAAAAGGA---GAAGAAAAAAUAAUGUUCAACAUUAUCGUUGACAUUUAUUAGC ------..((..--..))...........((((((....(---------(((.((.(....)...)).---..........(((((.....))))))))).....))))))... ( -10.00, z-score = 0.27, R) >droSec1.super_15 1796056 94 - 1954846 ------AAGCAG--AAGCAGAUAUACAAAGAUAGAGAAUG---------CGGGCCGUGAAAAAAACGA---GAAGAAAAAAUAAUGUUCAACAUUAUCGUUGACAUUUAUUAGC ------..((..--..(((..(((......))).....))---------)..))(((.......))).---........((((((((.((((......))))))).)))))... ( -11.50, z-score = -0.34, R) >droYak2.chrX 10734904 111 + 21770863 AAGCAGAAGCAGAUACACAGAUACACAGAGAUAGAGAGUGGGGGAGGGAGGGGGCGGUGAGAAAAGGA---GAAGAAAAAAUAAUGUUCAACAUUAUCGUUGACAUUUAUUAGC ........((.((((........(((...........))).........(..((((((((.....(..---(((.(........).)))..).))))))))..)...)))).)) ( -10.90, z-score = -0.23, R) >droEre2.scaffold_4690 14708962 92 + 18748788 ------AGGCAG--AGGCAGAUUCACUGGGAUAGCGAGU-----------GAGGCAGUGAAAAAAGGA---GAAGAAAAAAUAAUGUUCAACAUUAUCGUUGACAUUUAUUAGC ------..((..--..((....(((((.(.....).)))-----------)).)).............---........((((((((.((((......))))))).))))).)) ( -14.70, z-score = -0.56, R) >droAna3.scaffold_13248 2653528 95 - 4840945 --------------AAGUAUGUGUUGGGGAAUAAGAAGCAGC----UUGGAGUACA-AAAAAAAAUGGCAAGAAGAAAAAAUAAUGUCCAACAUUAUCGUUGACAUUUAUUAGC --------------.((((.((((..(.((...........(----(((..((...-.......))..)))).........(((((.....))))))).)..)))).))))... ( -13.30, z-score = -0.09, R) >dp4.chrXL_group1e 9191758 76 + 12523060 --------------------------------AAGAAGGAGA----ACAACAGCCACAAAAAAGGCGGCA--AAGAAAAAAUAAUGUCCAACAUUAUCGUUGACAUUUAUUAGC --------------------------------..........----.((((.(((........)))....--........((((((.....)))))).))))............ ( -11.40, z-score = -1.77, R) >droPer1.super_17 1662072 76 + 1930428 --------------------------------AAGAAGGAGA----ACAACAGCCACAAAAAAGGCGGCA--AAGAAAAAAUAAUGUCCAACAUUAUCGUUGACAUUUAUUAGC --------------------------------..........----.((((.(((........)))....--........((((((.....)))))).))))............ ( -11.40, z-score = -1.77, R) >droMoj3.scaffold_6473 10195715 78 - 16943266 -----------------------------ACCAGUGAGUCUAUGCUUUUAUGUAUGCACACACAC-------AUACAUACAUAAUGUUCAACAUUAUCGUGGACAUUUAUUAGC -----------------------------........(((((((....((((((((........)-------))))))).((((((.....))))))))))))).......... ( -17.90, z-score = -2.18, R) >consensus ______A_GCAG__AAGCAGAUA_ACAGAGAUAGAGAGUG_________AGGGCCGCAAAAAAAAGGA___GAAGAAAAAAUAAUGUUCAACAUUAUCGUUGACAUUUAUUAGC ..................................................................................(((((.((((......)))))))))....... ( -5.56 = -5.67 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:30 2011