| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,076,378 – 11,076,474 |
| Length | 96 |
| Max. P | 0.986972 |

| Location | 11,076,378 – 11,076,474 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Shannon entropy | 0.45481 |
| G+C content | 0.36720 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.64 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986972 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 11076378 96 + 22422827 ----------CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGC--AACAGCGG--GCGGCGAAUGAUC ----------((((((((.......(((((((((.(((((......))))).))))))))).......((((((...)))).(((--....))).--))))))))))... ( -33.90, z-score = -4.36, R) >droPer1.super_17 275738 93 + 1930428 ----------CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCAUGC--AACAGCAA--CCGAUUGAUC--- ----------...((((.(((((..(((((((((.(((((......))))).)))))))))..(((((....))))).)))))))--))......--..........--- ( -21.80, z-score = -1.27, R) >dp4.chrXL_group1e 3338576 93 + 12523060 ----------CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCAUGC--AACAGCAA--CCGAUUGAUC--- ----------...((((.(((((..(((((((((.(((((......))))).)))))))))..(((((....))))).)))))))--))......--..........--- ( -21.80, z-score = -1.27, R) >droAna3.scaffold_13248 2632886 97 + 4840945 ----------CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCAGAA--AACAACAAA-GCAGAGGGCGGGC ----------..((((((.(((...(((((((((.(((((......))))).))))))))).........((((...))))....--.........-)))...)))))). ( -24.90, z-score = -2.34, R) >droEre2.scaffold_4690 14693999 96 - 18748788 ----------CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGC--AACAGCAG--GCGGCAAAUGAUC ----------((((((((.......(((((((((.(((((......))))).))))))))).......((((((...))))..((--....))..--))))))))))... ( -32.00, z-score = -3.90, R) >droYak2.chrX 10719525 96 - 21770863 ----------CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGC--AACAGCAG--GCGGCAAGUGAUC ----------((((((((.......(((((((((.(((((......))))).))))))))).......((((((...))))..((--....))..--))))))))))... ( -32.00, z-score = -4.08, R) >droSec1.super_15 1779873 96 + 1954846 ----------CAUUUGCCGUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGC--AACAGCGG--GCGGCGAAUGAUC ----------((((((((((.(...(((((((((.(((((......))))).)))(((((((.....)))))))...))))))((--....))).--))))))))))... ( -35.30, z-score = -4.58, R) >droSim1.chrX 8574673 96 + 17042790 ----------CAUUUGCCGUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGC--AACAGCGG--GCGGCGAAUGAUC ----------((((((((((.(...(((((((((.(((((......))))).)))(((((((.....)))))))...))))))((--....))).--))))))))))... ( -35.30, z-score = -4.58, R) >droGri2.scaffold_15203 3112856 102 + 11997470 CAUCGAAACGCGUCAGGC-UGUAAAGUGCAUAAUAAAAUUGUUGAAA-UUUAAUUAUGCGCAAUUAAUGCGCAUUAAGUGCAGCA---ACAGGACGUUCAGCAAGUG--- ....((...(((((..(.-(((...(((((((((.(((((.....))-))).)))))))))......(((((.....))))))))---.)..)))))))........--- ( -29.30, z-score = -2.28, R) >droVir3.scaffold_12970 11135132 105 + 11907090 CGUCGCAGCGCGUCAGGC-UGUAAAGUGCAUAAUAAAAUUGUAGAAA-UUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCAGCA---ACAGGAUAUUCAGCGACUGAGU .(((((.....(((..((-((((..(((((((((.(((((.....))-))).)))))))))..(((((....))))).)))))).---....))).....)))))..... ( -32.10, z-score = -2.41, R) >droWil1.scaffold_180777 4630218 101 + 4753960 -CAUUGAACGCAUCCGAGCGCCACAUUGCAUAAUAAAAUUGUAAAAA-UUUAAUUAUGGGCAAUUAAUACGCAUUAA-UGCAGUC--AACCAAACG-ACACAUACUA--- -..((((..((((....(((....((((((((((.(((((.....))-))).)))))..))))).....)))....)-)))..))--)).......-..........--- ( -16.90, z-score = -0.81, R) >droMoj3.scaffold_6473 6716469 108 - 16943266 CAUCGGAGCGCGUCAGGC-UGUAAAGUGCAUAAUAAAAUUGUAGAAA-UUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCAGCAGCAACAGGAUAUCCAGCGAGUGAGU ..(((...(((((...((-(((...(((((((((.(((((.....))-))).))))))))).........((((...)))).))))).)).((....)).)))..))).. ( -31.20, z-score = -2.34, R) >consensus __________CAUUUGCCAUGUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGC__AACAGCAG__GCGGCGAAUGAUC ...................(((...((((((.....(((((.........)))))(((((((.....)))))))...)))))).....)))................... (-13.28 = -13.64 + 0.36)
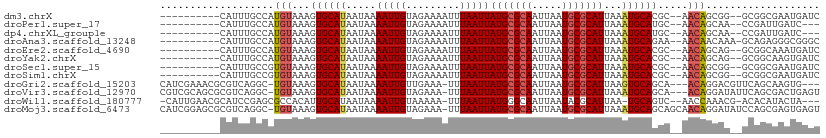
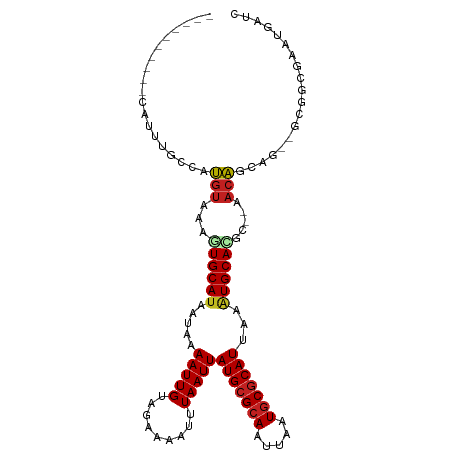
| Location | 11,076,378 – 11,076,474 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Shannon entropy | 0.45481 |
| G+C content | 0.36720 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -11.08 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859112 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 11076378 96 - 22422827 GAUCAUUCGCCGC--CCGCUGUU--GCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG---------- ...((((.((((.--..((....--))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))......)))).))))---------- ( -25.90, z-score = -2.57, R) >droPer1.super_17 275738 93 - 1930428 ---GAUCAAUCGG--UUGCUGUU--GCAUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG---------- ---..........--(((((((.--((((.......))))(((.....)))(((((((.(((((......))))).))))))).......)))))))...---------- ( -20.40, z-score = -0.92, R) >dp4.chrXL_group1e 3338576 93 - 12523060 ---GAUCAAUCGG--UUGCUGUU--GCAUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG---------- ---..........--(((((((.--((((.......))))(((.....)))(((((((.(((((......))))).))))))).......)))))))...---------- ( -20.40, z-score = -0.92, R) >droAna3.scaffold_13248 2632886 97 - 4840945 GCCCGCCCUCUGC-UUUGUUGUU--UUCUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG---------- ..........(((-(.(((.((.--....(((.(((((....))))).)))(((((((.(((((......))))).)))))))))...))).))))....---------- ( -20.50, z-score = -1.86, R) >droEre2.scaffold_4690 14693999 96 + 18748788 GAUCAUUUGCCGC--CUGCUGUU--GCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG---------- ...(((((((((.--..((....--))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))......)))))))))---------- ( -29.70, z-score = -3.52, R) >droYak2.chrX 10719525 96 + 21770863 GAUCACUUGCCGC--CUGCUGUU--GCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG---------- ...((.((((((.--..((....--))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))......)))))).))---------- ( -26.00, z-score = -2.71, R) >droSec1.super_15 1779873 96 - 1954846 GAUCAUUCGCCGC--CCGCUGUU--GCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACACGGCAAAUG---------- ...((((.((((.--..((....--))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))......)))).))))---------- ( -27.50, z-score = -3.28, R) >droSim1.chrX 8574673 96 - 17042790 GAUCAUUCGCCGC--CCGCUGUU--GCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACACGGCAAAUG---------- ...((((.((((.--..((....--))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))......)))).))))---------- ( -27.50, z-score = -3.28, R) >droGri2.scaffold_15203 3112856 102 - 11997470 ---CACUUGCUGAACGUCCUGU---UGCUGCACUUAAUGCGCAUUAAUUGCGCAUAAUUAAA-UUUCAACAAUUUUAUUAUGCACUUUACA-GCCUGACGCGUUUCGAUG ---.......((((((((..((---((..(((.(((((....))))).)))(((((((.(((-((.....))))).)))))))......))-))..))))...))))... ( -22.20, z-score = -1.41, R) >droVir3.scaffold_12970 11135132 105 - 11907090 ACUCAGUCGCUGAAUAUCCUGU---UGCUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAA-UUUCUACAAUUUUAUUAUGCACUUUACA-GCCUGACGCGCUGCGACG .....(((((.(....((..((---((..(((.(((((....))))).)))(((((((.(((-((.....))))).)))))))......))-))..))....).))))). ( -26.70, z-score = -2.28, R) >droWil1.scaffold_180777 4630218 101 - 4753960 ---UAGUAUGUGU-CGUUUGGUU--GACUGCA-UUAAUGCGUAUUAAUUGCCCAUAAUUAAA-UUUUUACAAUUUUAUUAUGCAAUGUGGCGCUCGGAUGCGUUCAAUG- ---..........-......(((--((.((((-((...((((....(((((..(((((.(((-((.....))))).))))))))))...))))...)))))).))))).- ( -25.60, z-score = -1.54, R) >droMoj3.scaffold_6473 6716469 108 + 16943266 ACUCACUCGCUGGAUAUCCUGUUGCUGCUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAA-UUUCUACAAUUUUAUUAUGCACUUUACA-GCCUGACGCGCUCCGAUG .......(((.((....)).(((((((..(((.(((((....))))).)))(((((((.(((-((.....))))).)))))))......))-))..))))))........ ( -24.20, z-score = -1.90, R) >consensus GAUCAUUCGCCGC__CUGCUGUU__GCCUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAUGGCAAAUG__________ ............................(((((...(((((((.....)))))))...((((...........))))..))))).......................... (-11.08 = -11.25 + 0.17)

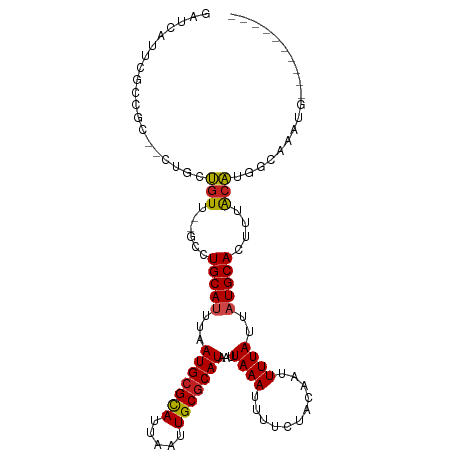
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:27 2011