| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,067,277 – 11,067,362 |
| Length | 85 |
| Max. P | 0.991430 |

| Location | 11,067,277 – 11,067,362 |
|---|---|
| Length | 85 |
| Sequences | 6 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Shannon entropy | 0.34610 |
| G+C content | 0.48451 |
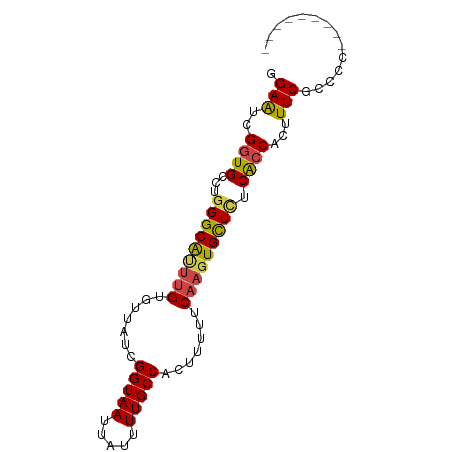
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
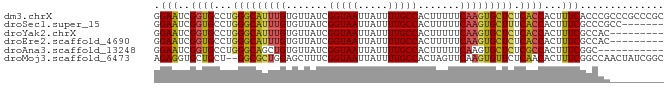
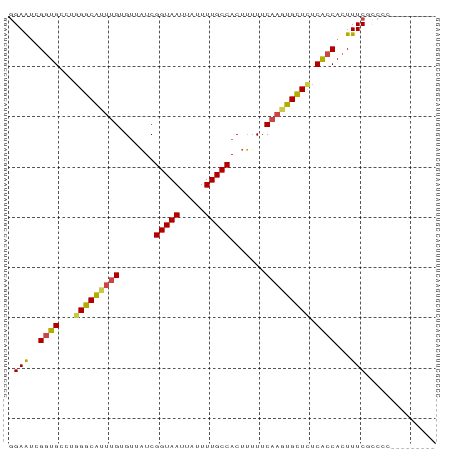
>dm3.chrX 11067277 85 - 22422827 GGAAUCGGUGCCUGGGCAUUUGUGUUAUCGGUAAUUAUUUUGCCACUUUUUCAAGUGCUCUCACCACUUUCACCCGCCCGCCCGC ((((..((((...(((((((((.......(((((.....))))).......))))))))).))))..)))).............. ( -24.04, z-score = -1.95, R) >droSec1.super_15 1770987 78 - 1954846 GGAAUCGGUGCCUGGGCAUUUGUGUUAUCGGUAAUUAUUUUGCCACUUUUUCAAGUGCUUUCACCACUUUCGCCCGCC------- ((((..((((...(((((((((.......(((((.....))))).......))))))))).))))..)))).......------- ( -22.34, z-score = -2.22, R) >droYak2.chrX 10710143 76 + 21770863 GGAAUCGGUGCCUGGGCAUUUGUGUUAUCGGUAAUUAUUUUGCCACUUUUUCAAGUGCUCUCACCACUUUCGCCAC--------- ((((..((((...(((((((((.......(((((.....))))).......))))))))).))))..)))).....--------- ( -24.04, z-score = -3.57, R) >droEre2.scaffold_4690 14684782 76 + 18748788 GGAAUCGGUGCCUGGGCAUUUGUGUUAUCGGUAAUUAUUUUGCCACUUUUUCAAGUGCUCUCACCACUUUCGCCAC--------- ((((..((((...(((((((((.......(((((.....))))).......))))))))).))))..)))).....--------- ( -24.04, z-score = -3.57, R) >droAna3.scaffold_13248 2614306 74 - 4840945 GGAAUCGGUGCCUGGGCAGCUGUGUUAUCGGUAAUUAUUUUGCCACUUUUUCAAGUGCUCUCGCCACUUUCGGC----------- ((((..((((...(((((..((.......(((((.....))))).......))..))))).))))..))))...----------- ( -18.84, z-score = -0.48, R) >droMoj3.scaffold_6473 6705112 83 + 16943266 AGAGGUGCUGCU--GGCGCUGGAGCUUUCGGUAAUUAUUUUGCCACUAGUUCAAGUGUUCUCAACACUUUCGGCCAACUAUCGGC .((((((.((..--((((((.(((((...(((((.....)))))...))))).))))))..)).))))))..(((.......))) ( -26.60, z-score = -1.84, R) >consensus GGAAUCGGUGCCUGGGCAUUUGUGUUAUCGGUAAUUAUUUUGCCACUUUUUCAAGUGCUCUCACCACUUUCGCCCC_________ .(((..((((...(((((((((.......(((((.....))))).......))))))))).))))...))).............. (-20.26 = -20.32 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:26 2011