| Sequence ID | dm3.chrX |
|---|---|
| Location | 11,039,759 – 11,039,879 |
| Length | 120 |
| Max. P | 0.610571 |

| Location | 11,039,759 – 11,039,879 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Shannon entropy | 0.35040 |
| G+C content | 0.51500 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 11039759 120 - 22422827 AAAUCGAACUACAACUUCGAGAAACCAUUCCUUUGGCUGGCGCGCAAGCUGGUUGGUGAUCCCAACCUGGAGUUUGUCGCCAUGCCAGCCCUGCUGCCGCCCGAGGUUAAGAUGGAUAAG ...(((((.......)))))....(((((((((.(((.((((.(((.((((((((((((((((.....)).)...))))))).))))))..)))))))))).))))....)))))..... ( -47.20, z-score = -3.71, R) >anoGam1.chr2L 48563539 120 - 48795086 AAAUCGAACUACAACUUCGAGAAACCGUUCCUGUGGCUGGCUCGCAAGCUUGUCGGUGAUCCUAACCUGGAGUUCGUUGCCAUGCCUGCCUUGCUUCCACCGGAAGUGAAAAUGGACAAG ...(((((.......)))))....(((((.....(((.(((..((((((((...(((.......)))..))))...))))...))).)))(..(((((...)))))..).)))))..... ( -32.90, z-score = -0.64, R) >droGri2.scaffold_14853 8498178 120 + 10151454 AAAUCGAAUUACAACUUUGAGAAACCCUUCCUUUGGCUCGCCCGCAAACUGGUCGGCGAUCCAAACCUGGAGUUUGUGGCAAUGCCGGCACUGCUGCCACCCGAGGUCAAAAUGGACAGA ...(((((.......)))))......(((((.((((.(((((.((......)).))))).))))((((((.((..(..(((.((....)).)))..).)))).))))......))).)). ( -33.40, z-score = -0.05, R) >droMoj3.scaffold_6473 238956 120 - 16943266 AAGUCGAAUUACAACUUUGAGAAACCAUUCCUUUGGCUGGCCCGCAAGCUGGUCGGUGAUCCAAAUCUGGAGUUCGUAGCAAUGCCGGCGCUGCUGCCACCGGAGGUCAAAAUGGAUAAA ...(((((.......)))))....((((((((((((.((((..(((.(((((((((...((((....))))..))).......))))))..))).)))))))))))....)))))..... ( -39.11, z-score = -1.12, R) >droVir3.scaffold_12928 2939348 120 + 7717345 AAAUCGAAUUACAACUUUGAGAAACCAUUCCUUUGGCUGGCCCGCAAACUGGUCGGCGAUCCAAAUCUGGAGUUCGUGGCAAUGCCGGCACUGCUGCCACCGGAGGUCAAAAUGGAUAAA ...(((((.......)))))....((((((((((((.((((..(((.....(((((((.((((....))))((.....))..)))))))..))).)))))))))))....)))))..... ( -38.20, z-score = -1.31, R) >droWil1.scaffold_181150 3326669 120 + 4952429 AAAUCGAAUUAUAACUUCGAAAAACCAUUCCUUUGGCUGGCCCGCAAGCUGGUCGGUGAUCCCAACUUAGAAUUUGUGGCUAUGCCAGCUCUUCUGCCGCCAGAGGUUAAAAUGGACAAA ...(((((.......)))))....(((((((((((((.(((.....(((((((..(((..(.(((........))).)..)))))))))).....)))))))))))....)))))..... ( -38.20, z-score = -2.67, R) >droPer1.super_17 87103 120 - 1930428 AAGUCAAACUACAAUUUCGAAAAACCAUUCCUCUGGCUGGCACGCAAAUUGGUUGGUGAUCCCAACCUAGAGUUCGUUGCCAUGCCGGCACUGCUGCCGCCAGAGGUGAAAAUGGACAAG ........................(((((((((((((.((((.(((....((((((.....))))))..........((((.....)))).)))))))))))))))....)))))..... ( -43.90, z-score = -3.51, R) >dp4.chrXL_group1e 3152187 120 - 12523060 AAGUCAAACUACAAUUUCGAAAAACCAUUCCUCUGGCUGGCACGCAAAUUGGUUGGUGAUCCCAACCUAGAGUUCGUUGCCAUGCCGGCACUGCUGCCGCCAGAGGUGAAAAUGGACAAG ........................(((((((((((((.((((.(((....((((((.....))))))..........((((.....)))).)))))))))))))))....)))))..... ( -43.90, z-score = -3.51, R) >droAna3.scaffold_13417 6099247 120 + 6960332 AAAUCGAAUUACAAUUUCGAGAAACCAUUCCUUUGGCUGGCGCGCAAGCUGGUCGGUGAUCCCAACCUGGAAUUCGUUGCCAUGCCAGCUCUACUGCCGCCGGAGGUUAAGAUGGACAAG ...(((((.......)))))....(((((((((((((.((((....(((((((.((..((.((.....)).....))..))..)))))))....))))))))))))....)))))..... ( -43.40, z-score = -2.72, R) >droEre2.scaffold_4690 14657853 120 + 18748788 AAAUCGAACUACAACUUCGAGAAACCAUUCCUCUGGCUGGCGCGCAAGCUGGUUGGUGAUCCCAACCUGGAGUUCGUCGCCAUGCCAGCCCUGCUGCCGCCCGAGGUCAAGAUGGACAAG ...(((((.......)))))....(((((((((.(((.((((.(((.((((((((((((((((.....)).)...))))))).))))))..)))))))))).))))....)))))..... ( -49.00, z-score = -3.66, R) >droYak2.chrX 10683596 120 + 21770863 AAAUCGAACUACAACUUCGAGAAACCAUUCCUGUGGCUGGCACGCAAGCUGGUUGGUGAUCCCAACCUGGAGUUCGUCGCCAUGCCAGCCCUGCUGCCGCCCGAGGUUAAGAUGGACAAG ...(((((.......)))))....((((((((..(((.((((.(((.((((((((((((((((.....)).)...))))))).))))))..))))))))))..)))....)))))..... ( -45.70, z-score = -3.08, R) >droSec1.super_15 1743365 120 - 1954846 AAAUCGAACUACAACUUCGAGAAACCAUUCCUCUGGCUGGCGCGCAAGCUGGUUGGUGAUCCCAACCUGGAGUUUGUCGCCAUGCCAGCCCUGCUGCCGCCCGAGGUCAAGAUGGAUAAG ...(((((.......)))))....(((((((((.(((.((((.(((.((((((((((((((((.....)).)...))))))).))))))..)))))))))).))))....)))))..... ( -49.00, z-score = -3.95, R) >droSim1.chrX_random 3097014 120 - 5698898 AAAUCGAACUACAACUUCGAGAAACCAUUCCUCUGGCUGGCGCGCAAGCUGGUUGGUGAUCCCAACCUGGAGUUUGUCGCCAUGCCAGCCCUGCUGCCGCCCGAGGUCAAGAUGGAUAAG ...(((((.......)))))....(((((((((.(((.((((.(((.((((((((((((((((.....)).)...))))))).))))))..)))))))))).))))....)))))..... ( -49.00, z-score = -3.95, R) >apiMel3.Group11 159803 120 - 12576330 AAAAGCAACUAUAAUUUUGAAAAACCUUUCUUAUGGUUGGCACGUAAACUGAUCGGUGAUCCAAAUCUCGAAUUUGUCGCAAUGCCAGCUCUUCUUCCACCAGAAGUUACAAUGGAUCCA ...(((((((((((....((((....))))))))))))((((((.........))((((..(((((.....)))))))))..)))).)))(((((......))))).............. ( -20.40, z-score = 0.41, R) >triCas2.ChLG9 1475710 120 + 15222296 AAAUCGAAUUACAACUUCGAAAAGCCGUUCCUGUGGUUGGCGCGCAAACUAAUCGGUGAUCCUAAUCUAGAGUUUGUGGCGAUGCCCGCACUGGUGCCCCCAGAAGUCACAAUGGACCCC ...(((((.......)))))....(((((..(((((.(.((.((((((((....(((.......)))...)))))))))).)...)))))((((.....)))).......)))))..... ( -30.00, z-score = 0.14, R) >consensus AAAUCGAACUACAACUUCGAGAAACCAUUCCUUUGGCUGGCGCGCAAGCUGGUCGGUGAUCCCAACCUGGAGUUCGUCGCCAUGCCAGCCCUGCUGCCGCCCGAGGUCAAAAUGGACAAG ...(((((.......)))))....(((((((((.((.((((.......((((((((...((........))..))).......))))).......)))))).))))....)))))..... (-19.24 = -19.08 + -0.15)

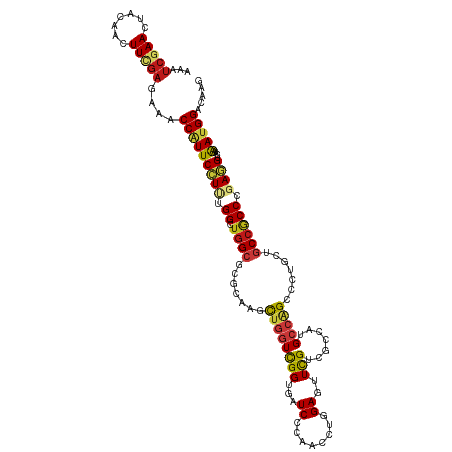
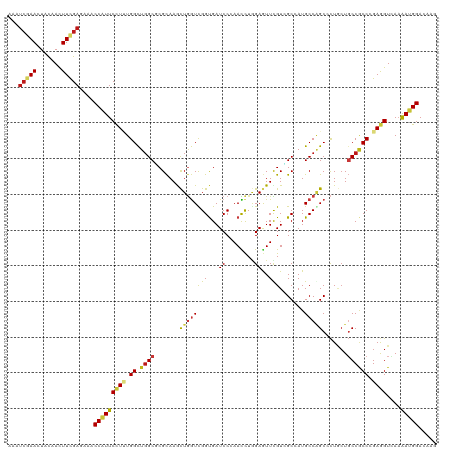
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:18 2011