
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,985,288 – 10,985,388 |
| Length | 100 |
| Max. P | 0.722144 |

| Location | 10,985,288 – 10,985,388 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 60.93 |
| Shannon entropy | 0.78200 |
| G+C content | 0.50660 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -10.60 |
| Energy contribution | -11.32 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.722144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
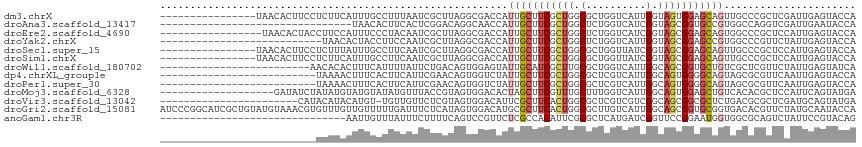
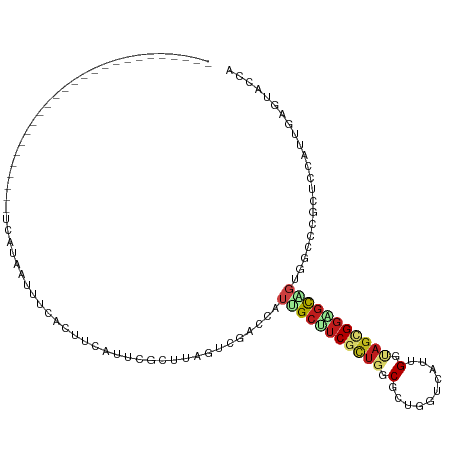
>dm3.chrX 10985288 100 + 22422827 ----------------UAACACUUCCUCUUCAUUUGCCUUUAAUCGCUUAGGCGACCAUUGCUUCGCUGGCGCUGGUCAUUGGUAGUGGAGCAGUUGCCCGCUCGAUUGAGUACCA ----------------......................((((((((....((((((...((((((((((.((........)).))))))))))))))))....))))))))..... ( -29.90, z-score = -0.96, R) >droAna3.scaffold_13417 6036514 84 - 6960332 --------------------------------UAACACUUCACUCGGACAGGCAACCAUUGCUUCGCUGGCUCUGGUCAUCGGUAGCGGUGCCGUGGCCAGGUCGAUUGAAUACCA --------------------------------......((((.(((.(((((((.....)))))..((((((.(((.(((((....)))))))).))))))))))).))))..... ( -29.70, z-score = -1.57, R) >droEre2.scaffold_4690 14607764 99 - 18748788 -----------------UAACACUACCUUCCAUUUCCCUACAAUCGCUUAGGCGACCAUUGCUUCGCUGGCUCUGGUCAUCGGUAGCGGAGCAGUGGCCCGCUCCAUUGAGUACCA -----------------............................(((((((((.((((((((((((((.(..........).))))))))))))))..))))....))))).... ( -31.60, z-score = -1.85, R) >droYak2.chrX 10637323 89 - 21770863 ---------------------------UAACACUACCUUCCAAUCGCUUAGGCGACCAUUGCUUCGCUGGCUCUGGUCAUUGGUAGCGGAGCCGUGGCCCGUUCUAUUGAGUACCA ---------------------------..................((((((..(((((..(((.....)))..)))))..(((.(((((.((....)))))))))))))))).... ( -26.80, z-score = -0.68, R) >droSec1.super_15 1696327 100 + 1954846 ----------------UAACACUUCCUCUUUAUUUGCCUUCAAUCGCUUAGGCGACCAUUGCUUCGCUGGCGCUGGUUAUCGGUAGCGGAGCAGUUGCCCGCUCCAUUGAGUACCA ----------------.........(((.......((((..........))))(((((.((((.....)))).)))))...((.(((((.((....)))))))))...)))..... ( -30.40, z-score = -1.26, R) >droSim1.chrX 8540509 100 + 17042790 ----------------UAACACUUCCUCUUCAUUUGCCUUCAAUCGCUUAGGCGACCAUUGCUUCGCUGGCGCUGGUUAUCGGUAGCGGAGCAGUUGCCCGCUCCAUUGAGUACCA ----------------.........(((.......((((..........))))(((((.((((.....)))).)))))...((.(((((.((....)))))))))...)))..... ( -30.40, z-score = -1.21, R) >droWil1.scaffold_180702 1044730 91 + 4511350 -------------------------AACACACUUUCAUUUUAUUCUGACAGUGGAGUAUUGCAUCGCUUGCGCUGGUCAUUGGCAGCGGUGCUGUCGCUCGUUCUAUUGAGUAUCA -------------------------(((.((((.(((........))).))))((((((.((((((((.((.(........))))))))))).)).)))))))............. ( -25.90, z-score = -1.04, R) >dp4.chrXL_group1e 899096 90 + 12523060 --------------------------UAAAACUUUCACUUCAUUCGAACAGUGGUCUAUUGCUUCGCUGGCGCUCGUCAUUGGCAGUGGGGCAGUAGCGCGUUCAAUUGAGUACCA --------------------------..........((((.(((.((((.(((..((((((((((((((.((........)).)))))))))))))))))))))))).)))).... ( -31.10, z-score = -2.90, R) >droPer1.super_30 654256 90 - 958394 --------------------------UAAAACUUUCACUUCAUUCGAACAGUGGUCUAUUGCUUCGCUGGCGCUCGUCAUUGGCAGUGGGGCAGUAGCGCGUUCAAUUGAGUACCA --------------------------..........((((.(((.((((.(((..((((((((((((((.((........)).)))))))))))))))))))))))).)))).... ( -31.10, z-score = -2.90, R) >droMoj3.scaffold_6328 899679 97 - 4453435 -------------------GAUAUCUAUAUGUAUGUAUAUGUUUACCGUAGUGGACACUAGCUUCGUUUGCUUUGGUCAUUGGCAGUGGAGCUGUCACACGCUCCAUUCAGUAUGA -------------------(((((.((((....)))).)))))...((((.(((((...(((.......)))...)))......((((((((........)))))))))).)))). ( -23.90, z-score = 0.05, R) >droVir3.scaffold_13042 2085753 92 - 5191987 -----------------------CAUACAUACAUGU-UGUUGUUCUCGUAGUGGACAUUCGCUUCACUGGCGCUCGUCGUCGGCAGCGGCGCUCUGACGCGCUCGAUGCAGUAUGA -----------------------(((((..((....-.))(((..(((.(((((((...((((.....))))...)))(((((.(((...)))))))).))))))).)))))))). ( -27.40, z-score = 0.96, R) >droGri2.scaffold_15081 3135778 116 + 4274704 AUCCCGGCAUCGCUGUAUGUAAACGUGUUUGUUGUUUUUGAUUUCUCAUAGUGGACAUGCGCUUCACUGGCGCUUGUCAUUGGCAGCGGUGCGGUGACACGUUCUAUGCAAUACCA ......((((((((((.....((((....))))...............(((((.(((.(((((.....))))).))))))))))))))))))((((....((.....))..)))). ( -35.60, z-score = -0.56, R) >anoGam1.chr3R 7843949 85 - 53272125 -------------------------------AAUUGUUUAUUUCUUUUCAGUCCGUUCUCGCCACAUUCGCGCUCAUGAUCGGUUCCGGAAUGGUGGCGCAGUCUAUUCCGUACAG -------------------------------.....................(((.((.(((.......))).....)).)))...((((((((.........))))))))..... ( -14.50, z-score = 0.77, R) >consensus __________________________UCAUAAUUUCACUUCAUUCGCUUAGUCGACCAUUGCUUCGCUGGCGCUGGUCAUUGGUAGCGGAGCAGUGGCCCGCUCCAUUGAGUACCA ..........................................................(((((((((((.(..........).)))))))))))...................... (-10.60 = -11.32 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:16 2011