
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,987,231 – 10,987,333 |
| Length | 102 |
| Max. P | 0.938316 |

| Location | 10,987,231 – 10,987,333 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 61.70 |
| Shannon entropy | 0.76711 |
| G+C content | 0.49598 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -11.30 |
| Energy contribution | -10.94 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10987231 102 + 23011544 --------AAUAAGAGUUUACAUACCGGCCCAUCACCACUACCUCGGGAAGUAGAGGAAUCAUC----UUCGU--UAAGUGGACGCCUGCGUCCACGUCCCUGAUCAUCAGCCAUU --------..................(((................((((....(((((....))----)))..--...(((((((....))))))).))))((.....)))))... ( -27.90, z-score = -1.84, R) >droEre2.scaffold_4929 12181656 102 - 26641161 --------GAAAUAAAUGUACAUACCGGUCCAUCACAACUUCCUCCGGAAUUAGAGGGCUCUUC----UUUGU--UAAGUGGGCGCCUGCGUCCACGUCCCUGAUCAUCAGCCAUA --------((..............((((................)))).(((((.((((.(...----...).--...(((((((....))))))))))))))))..))....... ( -24.29, z-score = -0.51, R) >droYak2.chr2L 7378905 108 + 22324452 --UAAAUCUGUUUGUGUCUACAUACCGGCCCAUCAUCACUUCCUCGGGAAUUGGAGGGUUCUUC----UUCGU--UAAGUGGACGCCUGCGUCCUCGACCCUGAUCAUCAGCCAUG --..........((((....))))..(((..((.((((.......(((.....(((((....))----)))..--...(.(((((....))))).)..))))))).))..)))... ( -24.41, z-score = 0.64, R) >droSec1.super_3 6388574 102 + 7220098 --------AAUAAGUGUUUACAUACCGGCCCAUCACCACUUCCUCGAGAAGUAGAGGAACCAUC----UUCGU--UAAGUGGACGCCUGCGUCCACGUCCCUGAUCAUCAGCCAUG --------.....(((....)))...(((..((((....((((((........)))))).....----.....--...(((((((....))))))).....)))).....)))... ( -26.80, z-score = -2.16, R) >droSim1.chr2L 10784354 102 + 22036055 --------AAUAAGUGUUAACAUACCGGUCCAUCACCACUUCCUCGAGGAGUAGAGGAAUCAUC----UUCGU--UAAGUGGACGCCUGCGUCCACGUCCCUGAUCAUCAGCCAUG --------........(((((.....(((.....)))..((((((........)))))).....----...))--)))(((((((....)))))))....(((.....)))..... ( -27.80, z-score = -1.80, R) >droWil1.scaffold_180708 4699943 108 + 12563649 --------AACAAAAUAUUACUUACCGGCUCUUCAGCUCGCUUUCGUCCUUCAUCAGAAUCAUCGGAAUUCCUGCUAUAUGGACGCCUGCGUCCGCGUUCCUGAUCAUCCGCCAUG --------..................(((...((((..((((((((...(((....)))....)))))............(((((....))))))))...))))......)))... ( -24.10, z-score = -1.77, R) >dp4.Unknown_group_260 92617 116 + 94298 AAAAUCUACAUACCGGUUCAUCUCUUUUUUCAUCAGUACUAGACCCUGGUCGGCCAAGAGAUUCAUCCAAUGUCCCAUGUGGGCGUCUAUGUCCACGUCCCUGAUUUUCCGACAUU ..............((...(((((((..(..(((((.........)))))..)..)))))))....))((((((..(((((((((....)))))))))....(......))))))) ( -25.00, z-score = -0.78, R) >droPer1.super_1 806949 116 + 10282868 AAAAUCUACAUACCGGUUCAUCUCUUUUUUCAUCAGUACUAGACCCUGGUCGGCCAAGAGAUUCAUCCAAUGUCCCAUGUGGGCGUCUAUGUCCACGUCCCUGAUUUUCCGACAUU ..............((...(((((((..(..(((((.........)))))..)..)))))))....))((((((..(((((((((....)))))))))....(......))))))) ( -25.00, z-score = -0.78, R) >droAna3.scaffold_12916 14848797 100 + 16180835 ----------UAGAAUAUCACCUACCGGCUCUUC--CCUUCCCUCAUGGGAGGUCG--AUGGUUGUUCAUCUU--CGAGUGGACGCCUGCGCCCUCGACCUUGAUCUUCGGCCAUA ----------...............(((((..((--((.........)))))))))--(((((((.(((...(--((((.((.((....)))))))))...)))....))))))). ( -32.30, z-score = -1.46, R) >consensus ________AAUAAGAGUUUACAUACCGGCCCAUCACCACUUCCUCGUGAAGUAGAGGAAUCAUC____UUCGU__UAAGUGGACGCCUGCGUCCACGUCCCUGAUCAUCAGCCAUG ..........................(((.................................................(((((((....)))))))......(.....).)))... (-11.30 = -10.94 + -0.36)

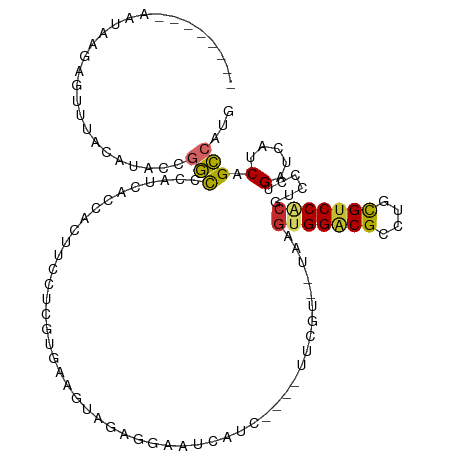
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:23 2011