| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,958,794 – 10,958,894 |
| Length | 100 |
| Max. P | 0.656441 |

| Location | 10,958,794 – 10,958,891 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 73.29 |
| Shannon entropy | 0.48131 |
| G+C content | 0.40137 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.12 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.649611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
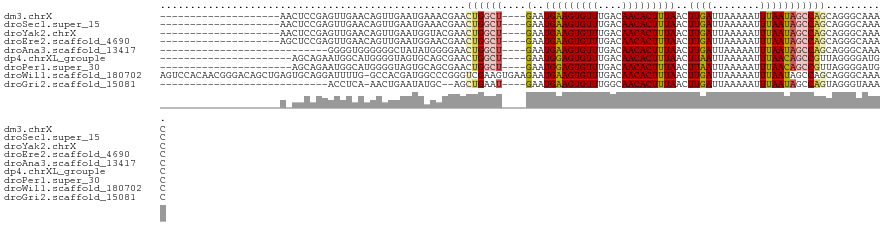
>dm3.chrX 10958794 97 + 22422827 --------------------AACUCCGAGUUGAACAGUUGAAUGAAACGAACUGGCU----GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC --------------------.........(((..(.(((......)))...((((((----(..(((((((((....)))))))))..((((........)))))))))))..)..))).. ( -21.30, z-score = -1.96, R) >droSec1.super_15 1670362 97 + 1954846 --------------------AACUCCGAGUUGAACAGUUGAAUGAAACGAACUGGCU----GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC --------------------.........(((..(.(((......)))...((((((----(..(((((((((....)))))))))..((((........)))))))))))..)..))).. ( -21.30, z-score = -1.96, R) >droYak2.chrX 10611995 97 - 21770863 --------------------AACUCCGAGUUGAACAGUUGAAUGGUACGAACUGGCU----GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC --------------------...(((..((....((......))..))...((((((----(..(((((((((....)))))))))..((((........)))))))))))..)))..... ( -19.80, z-score = -0.83, R) >droEre2.scaffold_4690 14582607 97 - 18748788 --------------------AGCUCCGAGUUGAACAGUUGAAUGGAACGAACUGGCU----GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC --------------------.((.((..(((...((......)).)))...((((((----(..(((((((((....)))))))))..((((........)))))))))))..)))).... ( -23.50, z-score = -2.03, R) >droAna3.scaffold_13417 6008101 89 - 6960332 ----------------------------GGGGUGGGGGGCUAUAUGGGGAACUGGCU----GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC ----------------------------..........(((...((.....((((((----(..(((((((((....)))))))))..((((........))))))))))))).))).... ( -20.10, z-score = -1.44, R) >dp4.chrXL_group1e 854280 95 + 12523060 ----------------------AGCAGAAUGGCAUGGGGUAGUGCAGCGAACUGGCU----GAAUGGAGUGUUUGACAACACUUUAACUUAAUUAAAAAUUUAACAGCCGUUAGGGGAUGC ----------------------.(((.((((((.((........((((......)))----)..(((((((((....)))))))))..................))))))))......))) ( -22.30, z-score = -0.78, R) >droPer1.super_30 610163 95 - 958394 ----------------------AGCAGAAUGGCAUGGGGUAGUGCAGCGAACUGGCU----GAAUGGAGUGUUUGACAACACUUUAACUUAAUUAAAAAUUUAACAGCCGUUAGGGGAUGC ----------------------.(((.((((((.((........((((......)))----)..(((((((((....)))))))))..................))))))))......))) ( -22.30, z-score = -0.78, R) >droWil1.scaffold_180702 3492970 120 - 4511350 AGUCCACAACGGGACAGCUGAGUGCAGGAUUUUG-GCCACGAUGGCCCGGGUCGAAGUGAAGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC .((((......)))).((((.((.((((((((((-(((.((......)))))))))))......(((((((((....))))))))).))))(((((....))))).))))))......... ( -35.10, z-score = -2.03, R) >droGri2.scaffold_15081 3103161 86 + 4274704 ----------------------------ACCUCA-AACUGAAUAUGC--AGCUGAAU----GAAUGAAGUGUUUGGCAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGUAGGGUAAAC ----------------------------((((..-.((((..(((..--.....(((----.(((((((((((....)))))))))..)).))).........)))..)))).)))).... ( -15.39, z-score = -0.68, R) >consensus ____________________A_CUCCGAGUUGAACAGUUGAAUGGAACGAACUGGCU____GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAAC ...................................................((((((.......(((((((((....)))))))))..((((........)))).)))))).......... (-11.80 = -12.12 + 0.32)

| Location | 10,958,794 – 10,958,891 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 73.29 |
| Shannon entropy | 0.48131 |
| G+C content | 0.40137 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -9.05 |
| Energy contribution | -9.43 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
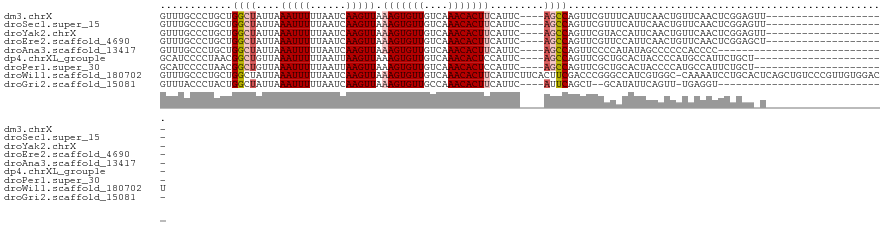
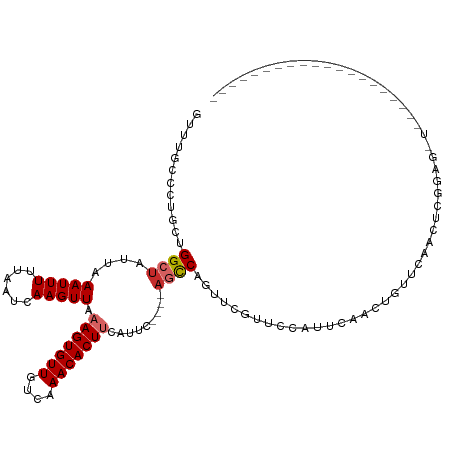
>dm3.chrX 10958794 97 - 22422827 GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC----AGCCAGUUCGUUUCAUUCAACUGUUCAACUCGGAGUU-------------------- ....((((.(((((((....(((((......))))).(((((((....))))))).....----)))))))..(((......)))..........)).)).-------------------- ( -17.60, z-score = -1.37, R) >droSec1.super_15 1670362 97 - 1954846 GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC----AGCCAGUUCGUUUCAUUCAACUGUUCAACUCGGAGUU-------------------- ....((((.(((((((....(((((......))))).(((((((....))))))).....----)))))))..(((......)))..........)).)).-------------------- ( -17.60, z-score = -1.37, R) >droYak2.chrX 10611995 97 + 21770863 GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC----AGCCAGUUCGUACCAUUCAACUGUUCAACUCGGAGUU-------------------- ...(((...(((((((....(((((......))))).(((((((....))))))).....----)))))))..)))........(((.......)))....-------------------- ( -18.60, z-score = -1.54, R) >droEre2.scaffold_4690 14582607 97 + 18748788 GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC----AGCCAGUUCGUUCCAUUCAACUGUUCAACUCGGAGCU-------------------- .........(((((((....(((((......))))).(((((((....))))))).....----)))))))..(((((.................))))).-------------------- ( -21.03, z-score = -2.47, R) >droAna3.scaffold_13417 6008101 89 + 6960332 GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC----AGCCAGUUCCCCAUAUAGCCCCCCACCCC---------------------------- ...((....(((((((....(((((......))))).(((((((....))))))).....----)))))))....))................---------------------------- ( -16.60, z-score = -3.37, R) >dp4.chrXL_group1e 854280 95 - 12523060 GCAUCCCCUAACGGCUGUUAAAUUUUUAAUUAAGUUAAAGUGUUGUCAAACACUCCAUUC----AGCCAGUUCGCUGCACUACCCCAUGCCAUUCUGCU---------------------- (((.........(((((...(((((......)))))..((((((....)))))).....)----))))........(((........))).....))).---------------------- ( -17.00, z-score = -1.69, R) >droPer1.super_30 610163 95 + 958394 GCAUCCCCUAACGGCUGUUAAAUUUUUAAUUAAGUUAAAGUGUUGUCAAACACUCCAUUC----AGCCAGUUCGCUGCACUACCCCAUGCCAUUCUGCU---------------------- (((.........(((((...(((((......)))))..((((((....)))))).....)----))))........(((........))).....))).---------------------- ( -17.00, z-score = -1.69, R) >droWil1.scaffold_180702 3492970 120 + 4511350 GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCUUCACUUCGACCCGGGCCAUCGUGGC-CAAAAUCCUGCACUCAGCUGUCCCGUUGUGGACU .........(((((.................(((...(((((((....)))))))....)))............((((.....)))-)............))))).((((......)))). ( -24.60, z-score = 0.01, R) >droGri2.scaffold_15081 3103161 86 - 4274704 GUUUACCCUACUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGCCAAACACUUCAUUC----AUUCAGCU--GCAUAUUCAGUU-UGAGGU---------------------------- ....(((....((((((((((....)))))..)))))(((((((....))))))).....----..((((((--(......))).)-))))))---------------------------- ( -12.50, z-score = 0.02, R) >consensus GUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC____AGCCAGUUCGUUCCAUUCAACUGUUCAACUCGGAG_U____________________ ............((((....(((((......))))).(((((((....))))))).........))))..................................................... ( -9.05 = -9.43 + 0.38)
| Location | 10,958,797 – 10,958,894 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 75.03 |
| Shannon entropy | 0.45779 |
| G+C content | 0.38587 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.08 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
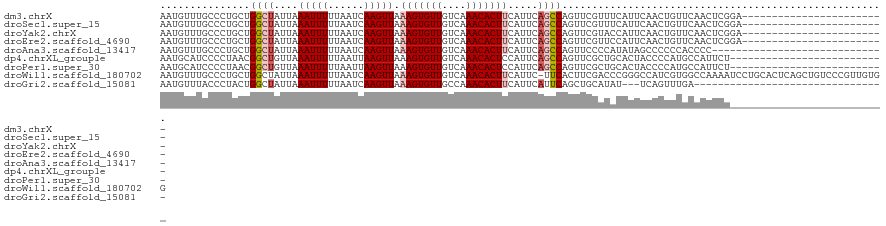
>dm3.chrX 10958797 97 + 22422827 ------------------------UCCGAGUUGAACAGUUGAAUGAAACGAACUGGCUGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU ------------------------......(((..(.(((......)))...(((((((..(((((((((....)))))))))..((((........)))))))))))..)..)))..... ( -21.30, z-score = -1.91, R) >droSec1.super_15 1670365 97 + 1954846 ------------------------UCCGAGUUGAACAGUUGAAUGAAACGAACUGGCUGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU ------------------------......(((..(.(((......)))...(((((((..(((((((((....)))))))))..((((........)))))))))))..)..)))..... ( -21.30, z-score = -1.91, R) >droYak2.chrX 10611998 97 - 21770863 ------------------------UCCGAGUUGAACAGUUGAAUGGUACGAACUGGCUGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU ------------------------.............(((.....(..(...(((((((..(((((((((....)))))))))..((((........)))))))))))..)..).)))... ( -19.90, z-score = -0.85, R) >droEre2.scaffold_4690 14582610 97 - 18748788 ------------------------UCCGAGUUGAACAGUUGAAUGGAACGAACUGGCUGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU ------------------------......(((..(.(((......)))...(((((((..(((((((((....)))))))))..((((........)))))))))))..)..)))..... ( -20.60, z-score = -1.37, R) >droAna3.scaffold_13417 6008101 92 - 6960332 -----------------------------GGGGUGGGGGGCUAUAUGGGGAACUGGCUGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU -----------------------------..((((....(((...((.....(((((((..(((((((((....)))))))))..((((........))))))))))))).)))...)))) ( -21.80, z-score = -1.65, R) >dp4.chrXL_group1e 854283 95 + 12523060 --------------------------AGAAUGGCAUGGGGUAGUGCAGCGAACUGGCUGAAUGGAGUGUUUGACAACACUUUAACUUAAUUAAAAAUUUAACAGCCGUUAGGGGAUGCAUU --------------------------......((((......)))).(((..(((((((..(((((((((....)))))))))..((((........))))))))).....))..)))... ( -22.20, z-score = -0.98, R) >droPer1.super_30 610166 95 - 958394 --------------------------AGAAUGGCAUGGGGUAGUGCAGCGAACUGGCUGAAUGGAGUGUUUGACAACACUUUAACUUAAUUAAAAAUUUAACAGCCGUUAGGGGAUGCAUU --------------------------......((((......)))).(((..(((((((..(((((((((....)))))))))..((((........))))))))).....))..)))... ( -22.20, z-score = -0.98, R) >droWil1.scaffold_180702 3492973 120 - 4511350 CCACAACGGGACAGCUGAGUGCAGGAUUUUGGCCACGAUGGCCCGGGUCGAAGUGAA-GAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU ((.....))....((((.((.(((((((((((((.((......)))))))))))...-...(((((((((....))))))))).))))(((((....))))).))))))............ ( -29.80, z-score = -0.71, R) >droGri2.scaffold_15081 3103164 86 + 4274704 --------------------------------UCAAACUGA---AUAUGCAGCUGAAUGAAUGAAGUGUUUGGCAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGUAGGGUAAACAUU --------------------------------.........---.(((.(.((((......(((((((((....))))))))).....(((((....)))))...)))).).)))...... ( -14.00, z-score = -0.22, R) >consensus __________________________CGAGUGGAACAGUGGAAUGGAACGAACUGGCUGAAUGAAGUGUUUGACAACACUUUAACUUGAUUAAAAAUUUAAUAGCCAGCAGGGCAAACAUU .....................................................((((((..(((((((((....)))))))))..((((........)))))))))).............. (-13.21 = -13.08 + -0.13)
| Location | 10,958,797 – 10,958,894 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Shannon entropy | 0.45779 |
| G+C content | 0.38587 |
| Mean single sequence MFE | -17.80 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
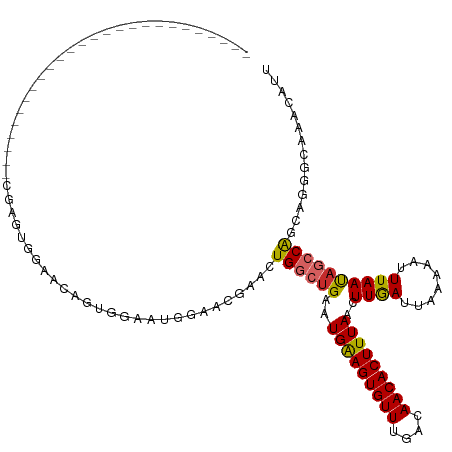
>dm3.chrX 10958797 97 - 22422827 AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCAGCCAGUUCGUUUCAUUCAACUGUUCAACUCGGA------------------------ ((((...((...(((((((....(((((......))))).(((((((....))))))).....)))))))..))..))))...(((.......))).------------------------ ( -19.50, z-score = -2.08, R) >droSec1.super_15 1670365 97 - 1954846 AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCAGCCAGUUCGUUUCAUUCAACUGUUCAACUCGGA------------------------ ((((...((...(((((((....(((((......))))).(((((((....))))))).....)))))))..))..))))...(((.......))).------------------------ ( -19.50, z-score = -2.08, R) >droYak2.chrX 10611998 97 + 21770863 AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCAGCCAGUUCGUACCAUUCAACUGUUCAACUCGGA------------------------ ((((..(((...(((((((....(((((......))))).(((((((....))))))).....)))))))..))).))))...(((.......))).------------------------ ( -19.10, z-score = -1.75, R) >droEre2.scaffold_4690 14582610 97 + 18748788 AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCAGCCAGUUCGUUCCAUUCAACUGUUCAACUCGGA------------------------ ((((...((...(((((((....(((((......))))).(((((((....))))))).....)))))))..))..))))...(((.......))).------------------------ ( -18.80, z-score = -1.86, R) >droAna3.scaffold_13417 6008101 92 + 6960332 AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCAGCCAGUUCCCCAUAUAGCCCCCCACCCC----------------------------- ..(((.((....(((((((....(((((......))))).(((((((....))))))).....)))))))....)).)))............----------------------------- ( -17.80, z-score = -3.42, R) >dp4.chrXL_group1e 854283 95 - 12523060 AAUGCAUCCCCUAACGGCUGUUAAAUUUUUAAUUAAGUUAAAGUGUUGUCAAACACUCCAUUCAGCCAGUUCGCUGCACUACCCCAUGCCAUUCU-------------------------- ..((((.(....((((((((...(((((......)))))..((((((....)))))).....))))).))).).)))).................-------------------------- ( -16.40, z-score = -1.81, R) >droPer1.super_30 610166 95 + 958394 AAUGCAUCCCCUAACGGCUGUUAAAUUUUUAAUUAAGUUAAAGUGUUGUCAAACACUCCAUUCAGCCAGUUCGCUGCACUACCCCAUGCCAUUCU-------------------------- ..((((.(....((((((((...(((((......)))))..((((((....)))))).....))))).))).).)))).................-------------------------- ( -16.40, z-score = -1.81, R) >droWil1.scaffold_180702 3492973 120 + 4511350 AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUC-UUCACUUCGACCCGGGCCAUCGUGGCCAAAAUCCUGCACUCAGCUGUCCCGUUGUGG .......(((.....)))................(((...(((((((....)))))))....)-))(((..((.....((((.....))))........((.....)).....))..))). ( -22.20, z-score = 0.46, R) >droGri2.scaffold_15081 3103164 86 - 4274704 AAUGUUUACCCUACUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGCCAAACACUUCAUUCAUUCAGCUGCAUAU---UCAGUUUGA-------------------------------- ..............((((((((((....)))))..)))))(((((((....))))))).......(((((((.....---.))).))))-------------------------------- ( -10.50, z-score = 0.31, R) >consensus AAUGUUUGCCCUGCUGGCUAUUAAAUUUUUAAUCAAGUUAAAGUGUUGUCAAACACUUCAUUCAGCCAGUUCGUUCCAUUCCACUGUGCAACUCG__________________________ ...............((((....(((((......))))).(((((((....))))))).....))))...................................................... ( -9.32 = -9.70 + 0.38)
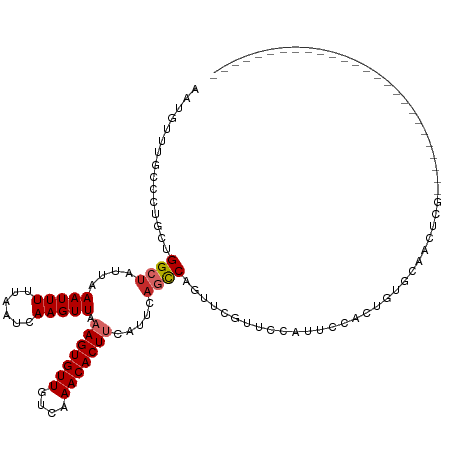
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:12 2011