| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,939,737 – 10,939,886 |
| Length | 149 |
| Max. P | 0.995884 |

| Location | 10,939,737 – 10,939,886 |
|---|---|
| Length | 149 |
| Sequences | 7 |
| Columns | 165 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.36801 |
| G+C content | 0.42814 |
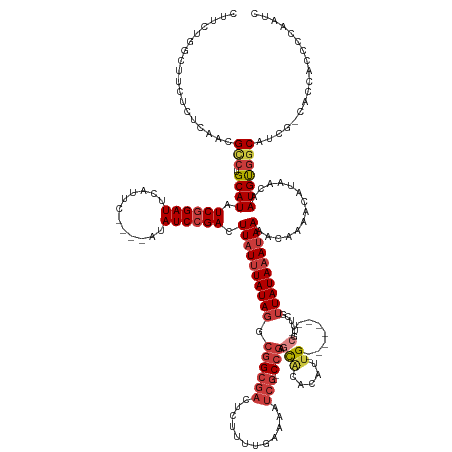
| Mean single sequence MFE | -49.19 |
| Consensus MFE | -28.84 |
| Energy contribution | -30.31 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
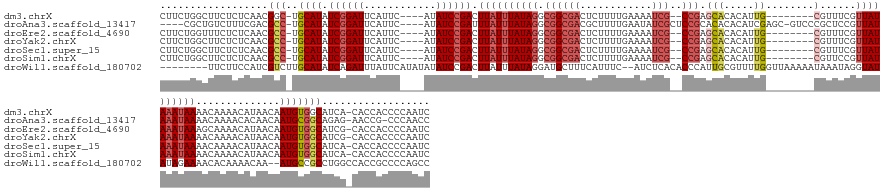
>dm3.chrX 10939737 149 + 22422827 GAUUGGGGUGGUG-UGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG--------CAAUGUGUGCUCGG--CGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAU----GAAUGAAUCCGAUAUGCA-GCCGUUGAGAGAAGCCAGAAG ..((((((((((.-.((.((((((((((...((((((((........)))))))))--------))))))).))))((--((((((......))))))))))).........(((((((..----......))))))).....-))).((....))..))))... ( -49.30, z-score = -3.76, R) >droAna3.scaffold_13417 5984378 153 - 6960332 GGUUGGG-CGGUU-CUCUGCCGCAUUGUUGUGUUUUGUUUUAUUUAUAACGGAGCGGGAC-GCUCGAUGUGUGUGCGGAGCGAUAUUCAAAAGCGUCGCCGCCUAUAAAUAAAUCGGAUAU----GAAUGAAUCCGAUAUGCA-GGCGUCGAAAGACAGCG---- ...((((-((((.-(((((((((((((..(((((((((((..(.....)..)))))))))-)).)))))))...)))))).(((..........))))))))))).......(((((((..----......))))))).....-.(((((....))).)).---- ( -55.70, z-score = -2.49, R) >droEre2.scaffold_4690 14564586 149 - 18748788 GAUUGGGGUGGUG-CGAUGCCACAUUGUUAUGUUUUGCUUUAUUUAUAACGAAACG--------CAAUGUGUGCUCGG--CGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAU----GAAUGAAUCCGAUAUGCA-GGCGUUGAGAGAAACCAGAAG ..((((.(((((.-....))))).((((((((..(......)..))))))))..(.--------((((((.(((.(((--((((((......)))))))))...........(((((((..----......)))))))..)))-.)))))).).....))))... ( -48.70, z-score = -3.21, R) >droYak2.chrX 10592795 149 - 21770863 GAUUGGGGUGGUG-CGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG--------CAAUGUGUGCUCGG--CGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAU----GAAUGAAUCCGAUAUGCA-GGCGUUGAGAGAAGCCAGAAG ..(((..((((.(-(((.((((((((((...((((((((........)))))))))--------))))))).))))((--((((((......))))))))))))))...)))(((((((..----......))))))).....-(((.((....)).)))..... ( -52.60, z-score = -4.29, R) >droSec1.super_15 1651506 149 + 1954846 GAUUGGGGUGGUG-UGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG--------CAAUGUGUGCUCGG--CGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAU----GAAUGAAUCCGAUAUGCA-GGCGUUGAGAGAAGCCAGAAG .(((.((((....-.((.((((((((((...((((((((........)))))))))--------))))))).))))((--((((((......))))))))))))...)))..(((((((..----......))))))).....-(((.((....)).)))..... ( -51.00, z-score = -4.14, R) >droSim1.chrX 8493875 149 + 17042790 GAUUGGGGUGGUG-UGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGGAACG--------CAAUGUGUGCUCGG--CGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAU----GAAUGAAUCCGAUAUGCA-GGCGUUGAGAGAAGCCAGAAG .(((.((((....-.((.((((((((((...((((((((........)))))))))--------))))))).))))((--((((((......))))))))))))...)))..(((((((..----......))))))).....-(((.((....)).)))..... ( -50.60, z-score = -3.79, R) >droWil1.scaffold_180702 996571 153 + 4511350 GGCUGGGGCGGUGGCCAGGCGGCAU--UUGUUUUGUGUUUUCUAUAUACCUAUUUAUUUUUAACCAAAACGCAAUGGGUGUGAGAU--GAAAUGAAAGCAUCCUAUAAAUAAGUCGGAUAUAUAUGAAUAAAUCUGAUAUGCAAGACGAUGGAAGAA-------- .((((.(((....)))...))))..--(((((((((((((((..((((((((((...((((....))))...))))))))))....--.....)))))).............(((((((.(((....))).)))))))..)))))))))........-------- ( -36.40, z-score = -0.55, R) >consensus GAUUGGGGUGGUG_CGAUGCCACAUUGUUAUGUUUUGUUUUAUUUAUAACGAAACG________CAAUGUGUGCUCGG__CGAUUUUCAAAAGAGUCGCCGCCUAUAAAUAAGUCGGAUAU____GAAUGAAUCCGAUAUGCA_GGCGUUGAGAGAAGCCAGAAG .....(((((((..(((.(((((((((...(((((((((........)))))))))........))))))).)))))....(((((......))))))))))))........(((((((............)))))))........................... (-28.84 = -30.31 + 1.48)
| Location | 10,939,737 – 10,939,886 |
|---|---|
| Length | 149 |
| Sequences | 7 |
| Columns | 165 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.36801 |
| G+C content | 0.42814 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -15.76 |
| Energy contribution | -15.46 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
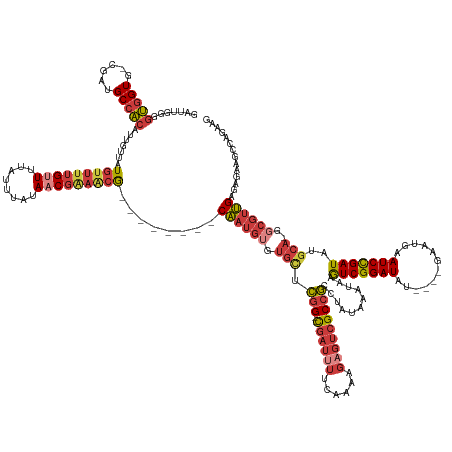
>dm3.chrX 10939737 149 - 22422827 CUUCUGGCUUCUCUCAACGGC-UGCAUAUCGGAUUCAUUC----AUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCG--CCGAGCACACAUUG--------CGUUUCGUUAUAAAUAAAACAAAACAUAACAAUGUGGCAUCA-CACCACCCCAAUC ....(((............((-..(((.((((((......----..)))))).((((((((((.((((((.((....))...)))--))).(((.....))--------)......))))))))))..............)))..))....-.......)))... ( -34.60, z-score = -2.80, R) >droAna3.scaffold_13417 5984378 153 + 6960332 ----CGCUGUCUUUCGACGCC-UGCAUAUCGGAUUCAUUC----AUAUCCGAUUUAUUUAUAGGCGGCGACGCUUUUGAAUAUCGCUCCGCACACACAUCGAGC-GUCCCGCUCCGUUAUAAAUAAAACAAAACACAACAAUGCGGCAGAG-AACCG-CCCAACC ----.((.((.((((...(((-.((((.((((((......----..))))))(((((((((((((((.(((((((.........((...)).........))))-))))))))....)))))))))).............))))))).)))-))).)-)...... ( -45.17, z-score = -3.66, R) >droEre2.scaffold_4690 14564586 149 + 18748788 CUUCUGGUUUCUCUCAACGCC-UGCAUAUCGGAUUCAUUC----AUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCG--CCGAGCACACAUUG--------CGUUUCGUUAUAAAUAAAGCAAAACAUAACAAUGUGGCAUCG-CACCACCCCAAUC ....((((..........(((-((.(((((((((......----..))))))..)))...)))))(((((.((....))...)))--))((((.(((((((--------.((((.(((........))).))))....))))))))).)).-.))))........ ( -37.60, z-score = -3.11, R) >droYak2.chrX 10592795 149 + 21770863 CUUCUGGCUUCUCUCAACGCC-UGCAUAUCGGAUUCAUUC----AUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCG--CCGAGCACACAUUG--------CGUUUCGUUAUAAAUAAAACAAAACAUAACAAUGUGGCAUCG-CACCACCCCAAUC ......((..........(((-((.(((((((((......----..))))))..)))...)))))(((((.((....))...)))--))((((.(((((((--------.((((.(((........))).))))....))))))))).)))-)............ ( -36.60, z-score = -3.29, R) >droSec1.super_15 1651506 149 - 1954846 CUUCUGGCUUCUCUCAACGCC-UGCAUAUCGGAUUCAUUC----AUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCG--CCGAGCACACAUUG--------CGUUUCGUUAUAAAUAAAACAAAACAUAACAAUGUGGCAUCA-CACCACCCCAAUC ....(((...........(((-((.(((((((((......----..))))))..)))...)))))(((((.((....))...)))--))((((.(((((((--------.((((.(((........))).))))....))))))))).)).-.......)))... ( -36.50, z-score = -3.79, R) >droSim1.chrX 8493875 149 - 17042790 CUUCUGGCUUCUCUCAACGCC-UGCAUAUCGGAUUCAUUC----AUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCG--CCGAGCACACAUUG--------CGUUCCGUUAUAAAUAAAACAAAACAUAACAAUGUGGCAUCA-CACCACCCCAAUC ....(((...........(((-((.(((((((((......----..))))))..)))...)))))(((((.((....))...)))--))((((.(((((((--------.(((..(((........)))..)))....))))))))).)).-.......)))... ( -34.80, z-score = -3.19, R) >droWil1.scaffold_180702 996571 153 - 4511350 --------UUCUUCCAUCGUCUUGCAUAUCAGAUUUAUUCAUAUAUAUCCGACUUAUUUAUAGGAUGCUUUCAUUUC--AUCUCACACCCAUUGCGUUUUGGUUAAAAAUAAAUAGGUAUAUAGAAAACACAAAACAA--AUGCCGCCUGGCCACCGCCCCAGCC --------...............((((....(((.(((....))).)))..((((((((((.(((((.........)--)))).....(((........)))......))))))))))....................--)))).((..(((....)))...)). ( -19.60, z-score = 1.11, R) >consensus CUUCUGGCUUCUCUCAACGCC_UGCAUAUCGGAUUCAUUC____AUAUCCGACUUAUUUAUAGGCGGCGACUCUUUUGAAAAUCG__CCGAGCACACAUUG________CGUUUCGUUAUAAAUAAAACAAAACAUAACAAUGUGGCAUCG_CACCACCCCAAUC ..................(((..((((.((((((............)))))).((((((((((.(((.(((.......................................))))))))))))))))..............))))))).................. (-15.76 = -15.46 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:04 2011