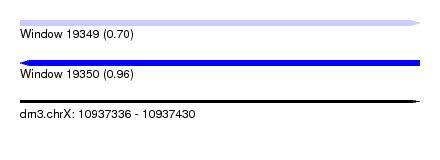
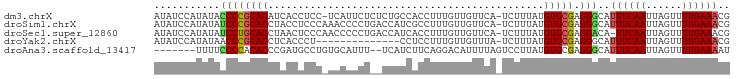
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,937,336 – 10,937,430 |
| Length | 94 |
| Max. P | 0.958619 |

| Location | 10,937,336 – 10,937,430 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.40 |
| Shannon entropy | 0.52928 |
| G+C content | 0.41845 |
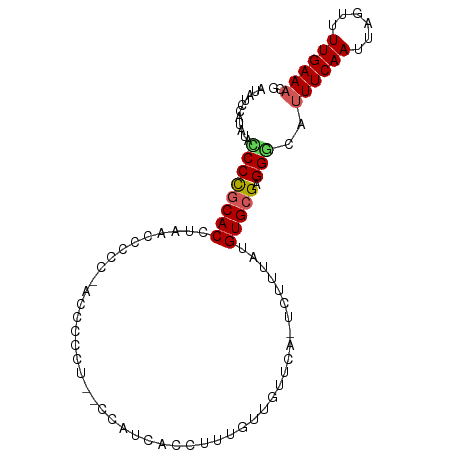
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.39 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
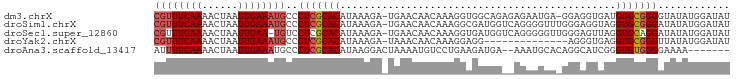
>dm3.chrX 10937336 94 + 22422827 CGUUUCAAAACUAAUUGAAAUGCCCUCGCACAUAAAGA-UGAACAACAAAGGUGGCAGAGAGAAUGA-GGAGGUGAUGUGCGGGGUAUAUGGAUAU ((((((((......)))))))).((((((((((.....-.......((....)).((.......)).-.......))))))))))........... ( -19.60, z-score = -1.01, R) >droSim1.chrX 8491437 95 + 17042790 CGUUUCAAAACUAAUUGAAAUGCCCUCGCACAUAAAGA-UGAACAACAAAGGCGAUGGUCAGGGGUUUGGGAGGUAGGUGCGGGAUAUAUGGAUAU ((((((((......))))))))..(((((((.((....-(((((..(...(((....))).)..))))).....)).)))))))............ ( -21.50, z-score = -0.97, R) >droSec1.super_12860 2 94 + 1111 CGUUUCAAAACUAAUUGAA-UGUCCUCGCACAUAAAGA-UGAACAACAAAGGUGAUGGUCAGGGGGUUGGGAGUUAGGUGCAGGAUAUAUGGAUAU ....((((......))))(-((((((.((((.(((...-(.(((..(...((......))..)..))).)...))).)))))))))))........ ( -18.90, z-score = -0.54, R) >droYak2.chrX 10590437 81 - 21770863 CGUUUCAAAACUAAUUGAAAUGCCCUCGCACAUAAAGA-UAAACAACAAAGGAGG--------------AGGGUGAGGUGCGGGUUAUAUGGAUAU ((((((((......))))))))..(((((((.......-....(......)....--------------........)))))))............ ( -14.30, z-score = -0.32, R) >droAna3.scaffold_13417 5982186 87 - 6960332 AUUUUCAAAACUAAUUGAAAUGCCCUCGCACAUAAGGACUAAAAUGUCCUGAAGAUGA--AAAUGCACAGGCAUCGGGUGUGGGGAAAA------- ..((((((......))))))...((((((((...(((((......)))))........--..((((....))))...))))))))....------- ( -25.40, z-score = -2.95, R) >consensus CGUUUCAAAACUAAUUGAAAUGCCCUCGCACAUAAAGA_UGAACAACAAAGGAGAUGG__AGAGGGA_GGGAGUGAGGUGCGGGAUAUAUGGAUAU ((((((((......))))))))..(((((((..............................................)))))))............ (-10.75 = -11.39 + 0.64)
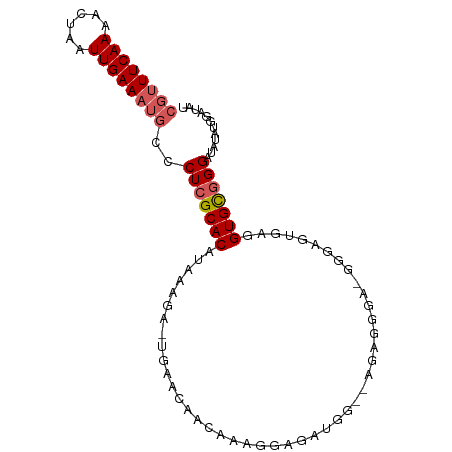
| Location | 10,937,336 – 10,937,430 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 71.40 |
| Shannon entropy | 0.52928 |
| G+C content | 0.41845 |
| Mean single sequence MFE | -15.19 |
| Consensus MFE | -9.31 |
| Energy contribution | -9.47 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 10937336 94 - 22422827 AUAUCCAUAUACCCCGCACAUCACCUCC-UCAUUCUCUCUGCCACCUUUGUUGUUCA-UCUUUAUGUGCGAGGGCAUUUCAAUUAGUUUUGAAACG ...........((((((((((.......-.((...................))....-.....))))))).)))..((((((......)))))).. ( -15.94, z-score = -2.04, R) >droSim1.chrX 8491437 95 - 17042790 AUAUCCAUAUAUCCCGCACCUACCUCCCAAACCCCUGACCAUCGCCUUUGUUGUUCA-UCUUUAUGUGCGAGGGCAUUUCAAUUAGUUUUGAAACG ...........................................((((((((....((-(....))).)))))))).((((((......)))))).. ( -14.80, z-score = -1.20, R) >droSec1.super_12860 2 94 - 1111 AUAUCCAUAUAUCCUGCACCUAACUCCCAACCCCCUGACCAUCACCUUUGUUGUUCA-UCUUUAUGUGCGAGGACA-UUCAAUUAGUUUUGAAACG ...........((((((((.(((....((((....((.....)).....))))....-...))).)))).))))..-(((((......)))))... ( -13.20, z-score = -1.39, R) >droYak2.chrX 10590437 81 + 21770863 AUAUCCAUAUAACCCGCACCUCACCCU--------------CCUCCUUUGUUGUUUA-UCUUUAUGUGCGAGGGCAUUUCAAUUAGUUUUGAAACG ............(((((((........--------------................-.......))))).))...((((((......)))))).. ( -10.98, z-score = -0.73, R) >droAna3.scaffold_13417 5982186 87 + 6960332 -------UUUUCCCCACACCCGAUGCCUGUGCAUUU--UCAUCUUCAGGACAUUUUAGUCCUUAUGUGCGAGGGCAUUUCAAUUAGUUUUGAAAAU -------..............(((((((.(((((..--........(((((......)))))...))))).)))))))((((......)))).... ( -21.02, z-score = -2.27, R) >consensus AUAUCCAUAUACCCCGCACCUAACCCCC_ACCCCCU__CCAUCACCUUUGUUGUUCA_UCUUUAUGUGCGAGGGCAUUUCAAUUAGUUUUGAAACG ...........((((((((..............................................))))).)))..((((((......)))))).. ( -9.31 = -9.47 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:33:02 2011