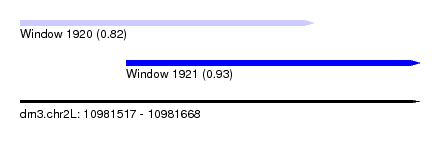
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,981,517 – 10,981,668 |
| Length | 151 |
| Max. P | 0.927148 |

| Location | 10,981,517 – 10,981,628 |
|---|---|
| Length | 111 |
| Sequences | 15 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Shannon entropy | 0.58224 |
| G+C content | 0.51946 |
| Mean single sequence MFE | -36.51 |
| Consensus MFE | -13.94 |
| Energy contribution | -14.21 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10981517 111 + 23011544 GCAAAAGGAAAUUGAACGAAACGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAG ((..............(((.....((.(((((.....))))).)).....((((((......)))))))))...((((((((((...---.))))))))))))........... ( -34.70, z-score = -1.28, R) >droSim1.chr2L 10778437 111 + 22036055 GCAAAAGGAAAUUGAACGAAACGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAG ((..............(((.....((.(((((.....))))).)).....((((((......)))))))))...((((((((((...---.))))))))))))........... ( -34.70, z-score = -1.28, R) >droSec1.super_3 6382497 111 + 7220098 GCAAAAGGAAAUUGAACGAAACGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAG ((..............(((.....((.(((((.....))))).)).....((((((......)))))))))...((((((((((...---.))))))))))))........... ( -34.70, z-score = -1.28, R) >droYak2.chr2L 7373700 111 + 22324452 GCAGAAAGAAAUAGAACGAAAUGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUUGAACUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAG (((....(........)....)))............(((.(((((..((.((((((......))))))..)).(((((((((((...---.))))))))))).)))).).))). ( -35.50, z-score = -2.13, R) >droEre2.scaffold_4929 12173802 111 - 26641161 GCAGAAGGAAAUUGAACGAAACGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUUGAAUUAUCCGAGUCUGA---GGAUUCGGAUAGCGAGGACGACAG .....................(((...(((((.....)))))((((((...(((((......))))))))))).((((((((((...---.))))))))))))).......... ( -34.30, z-score = -1.28, R) >droAna3.scaffold_12916 14843839 111 + 16180835 GCAGAAGGAAUUUGAACGUAAUACGGCACCUUUGGACGUCGCCUCCAUCUUGGCCAGACGCGUGGCAAUUGAACUGUCCGAGUCGGA---GGACUCGGACAGCGAUGACGACAG ((.(((((....((..((.....)).)))))))....((((((........)))..)))))((.(..((((..(((((((((((...---.)))))))))))))))..).)).. ( -37.50, z-score = -1.36, R) >dp4.Unknown_group_260 84005 111 + 94298 GCAGAAGGAGAUCGAACGCAACACAGCACCACUUGAUGUGGCAUCGAUCUUGGCAAGGCGUGUGGCGAUUGAGUUGUCCGAGUCUGA---AGAUUCGGAUAGUGAAGACGACAG ((.....((((((((..((......)).((((.....))))..))))))))......)).(((.(...((.(.(((((((((((...---.)))))))))))).))..).))). ( -35.70, z-score = -1.25, R) >droPer1.super_1 799446 111 + 10282868 GCAGAAGGAGAUCGAACGCAACACAGCACCACUUGAUGUGGCAUCGAUCUUGGCAAGACGUGUGGCGAUUGAGUUGUCCGAGUCUGA---AGAUUCGGAUAGUGAAGACGACAG ((.....((((((((..((......)).((((.....))))..)))))))).))......(((.(...((.(.(((((((((((...---.)))))))))))).))..).))). ( -34.80, z-score = -1.24, R) >droWil1.scaffold_180708 7876256 111 - 12563649 GCAAAAGGAAAUUGAACGUAAUACGGCACUUUUGGAUGUGGCAUCUAUAUUGGCAAGACGGGUGGCAAUUGAGUUAUCCGAAUCAGA---GGAUUCGGAUAGUGAAGAUGAUAG .........(((((..(((......((.....((((((...)))))).....))...))).....)))))...(((((((((((...---.)))))))))))............ ( -25.20, z-score = -1.19, R) >droVir3.scaffold_12963 19292171 111 - 20206255 GCUGAAGGAAAUUGAACGCAAUACGGCAAUGCUGGAUGUAGCCUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUAUCGGAGUCAGA---AGAUUCGGAUAGCGAGGAUGACAG ..........((((....)))).((((...))))..(((..((((.....((((((......))))))....((((((.(((((...---.))))).))))))))))...))). ( -38.20, z-score = -2.60, R) >droMoj3.scaffold_6500 4728745 111 - 32352404 GUUGAAGGAAAUUGAACGCAAUACGGCAAUGUUGGAUGUGGCAUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUGUCGGAGUCGGA---AGAUUCCGACAGUGAAGAUGACAG (((....((((((((.((((...((((...))))..))))...))))))))(((((......)))))(((..((((((((((((...---.))))))))))))...)))))).. ( -44.40, z-score = -4.81, R) >droGri2.scaffold_15252 15129979 111 - 17193109 GCUAAAGGAAAUUGAACGUAAUACGGCAAUGCUGGAUGUGGCCUCAAUAUUGGCCAGGCGAGUGGCCAUCGAGCUGUCGGAGUCGGA---GGACUCCGAUAGUGAAGAUGACAG (((.......((((..((.....)).))))(((.(.(.(((((........))))).)).))))))((((..((((((((((((...---.))))))))))))...)))).... ( -43.70, z-score = -3.99, R) >anoGam1.chr3R 11747961 111 + 53272125 GGCGAAGGAGAACGAUCGCAACAAGGGCCUGCACGAUGUGGCCUCCAUCCUUGCCCGUCGCGUCGCCAUCGAGCUGUCCGAGUCGGA---GUCGUCCGAAAGCGACGACGACAG (((.(((((((....)).......(((((.((.....)))))))...)))))))).((((((((((..(((..(..((((...))))---...)..)))..)))))).)))).. ( -43.50, z-score = -0.83, R) >apiMel3.Group3 1666364 114 + 12341916 GCAAAAGGAAGUGGAACGCGUGAACGCGUUGAACGACGUUGCAUCCAUAUUGGCGCGGCGAGUUGCUGUAGAAUUUAGCGACAGUGACUCAGCAUCGGAGAGCGAAUGUGACAG ((((......((..((((((....))))))..))....))))...((((((.((.(.((((((..((((.(.......).))))..)))).))......).)).)))))).... ( -39.50, z-score = -1.86, R) >triCas2.ChLG8 1963089 114 + 15773733 ACAGAAGGAGGUGGAGAGAGGGAACGGUUUCCACGAUGUUGCGAGUAUUUUAGCUCGGAGGGUUGCAGUCGAGUUUAGCGAUUCGGAUUCAGGAAGCGAUUCGGAGUGUGAUAG ..........(((((((..(....)..)))))))..(((..(((((......))))...(((((((..((((((((........)))))).))..))))))).....)..))). ( -31.30, z-score = -1.32, R) >consensus GCAGAAGGAAAUUGAACGAAACACGGCACCAUUGGAUGUGGCAUCAAUCUUGGCCAGACGUGUGGCUAUUGAGUUAUCCGAGUCGGA___GGAUUCGGAUAGCGAAGACGACAG ....................................(((.((.((.......((..........)).......(((((((((((.......))))))))))).)).).).))). (-13.94 = -14.21 + 0.27)
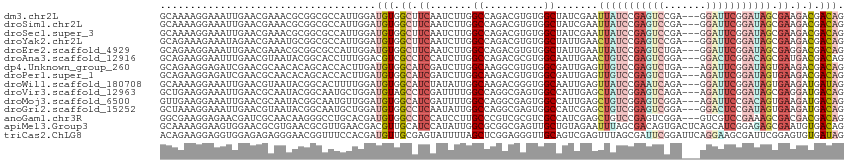

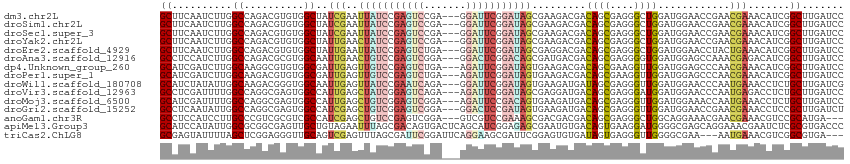
| Location | 10,981,557 – 10,981,668 |
|---|---|
| Length | 111 |
| Sequences | 15 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Shannon entropy | 0.53396 |
| G+C content | 0.54463 |
| Mean single sequence MFE | -40.31 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.17 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10981557 111 + 23011544 GCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACAUCGGCUUGAUCC ...........((((...(((((.(((.(((...((((((((((...---.)))))))))).))))).).))).))..))))((((.((.((((........)))).)).)))) ( -40.20, z-score = -2.12, R) >droSim1.chr2L 10778477 111 + 22036055 GCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACAUCGGCUUGAUCC ...........((((...(((((.(((.(((...((((((((((...---.)))))))))).))))).).))).))..))))((((.((.((((........)))).)).)))) ( -40.20, z-score = -2.12, R) >droSec1.super_3 6382537 111 + 7220098 GCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACAUCGGCUUGAUCC ...........((((...(((((.(((.(((...((((((((((...---.)))))))))).))))).).))).))..))))((((.((.((((........)))).)).)))) ( -40.20, z-score = -2.12, R) >droYak2.chr2L 7373740 111 + 22324452 GCUUCAAUCUUGGCCAGACGUGUGGCUAUUGAACUAUCCGAGUCCGA---GGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACAUCGGCUUGAUCC .((((..((.((((((......))))))..)).(((((((((((...---.))))))))))).))))....((((....))))(((.((.((((........)))).)).))). ( -42.50, z-score = -3.10, R) >droEre2.scaffold_4929 12173842 111 - 26641161 GCUUCAAUCUUGGCCAGACGUGUGGCUAUUGAAUUAUCCGAGUCUGA---GGAUUCGGAUAGCGAGGACGACAGCGAGGGCUGGAUGGAACCUACUGAAACAUCGGCUUGAUCC ((((((((...(((((......))))))))))).((((((((((...---.))))))))))))..((((((((((....))))((((.............))))...))).))) ( -35.82, z-score = -1.25, R) >droAna3.scaffold_12916 14843879 111 + 16180835 GCCUCCAUCUUGGCCAGACGCGUGGCAAUUGAACUGUCCGAGUCGGA---GGACUCGGACAGCGAUGACGACAGCGAGGGGUGGAUGGAGCCAAACGAGACAUCGGCUUGAUCC ..((((((((..(((...(((((.(..((((..(((((((((((...---.)))))))))))))))..).)).)))...))))))))))).(((.(((....)))..))).... ( -48.00, z-score = -2.94, R) >dp4.Unknown_group_260 84045 111 + 94298 GCAUCGAUCUUGGCAAGGCGUGUGGCGAUUGAGUUGUCCGAGUCUGA---AGAUUCGGAUAGUGAAGACGACAGCGAAGGUUGGAUGGAGCCCAACGAAACAUCGGCUUGAUCC ...(((.((((.((...((.....))........((((((((((...---.)))))))))))).)))))))((((....))))(((.(((((............))))).))). ( -38.60, z-score = -1.73, R) >droPer1.super_1 799486 111 + 10282868 GCAUCGAUCUUGGCAAGACGUGUGGCGAUUGAGUUGUCCGAGUCUGA---AGAUUCGGAUAGUGAAGACGACAGCGAAGGUUGGAUGGAGCCCAACGAAACAUCGGCUUGAUCC ...(((.((((.((....((.....)).......((((((((((...---.)))))))))))).)))))))((((....))))(((.(((((............))))).))). ( -38.10, z-score = -1.85, R) >droWil1.scaffold_180708 7876296 111 - 12563649 GCAUCUAUAUUGGCAAGACGGGUGGCAAUUGAGUUAUCCGAAUCAGA---GGAUUCGGAUAGUGAAGAUGAUAGCGAGGGUUGGAUGGAACCCAAUGAAACCUCUGCUUGAUCG .(((((..((((.((.......)).))))....(((((((((((...---.)))))))))))...)))))..((((((((((((.......)))))....)))).)))...... ( -32.30, z-score = -1.35, R) >droVir3.scaffold_12963 19292211 111 - 20206255 GCCUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUAUCGGAGUCAGA---AGAUUCGGAUAGCGAGGAUGACAGCGAGGGAUGGAUGGAACCCAAUGAGACCUCUGCUUGAUCC .((((.....((((((......))))))....((((((.(((((...---.))))).)))))))))).....(((((((..(.(.(((...))).).)..)))).)))...... ( -43.40, z-score = -2.62, R) >droMoj3.scaffold_6500 4728785 111 - 32352404 GCAUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUGUCGGAGUCGGA---AGAUUCCGACAGUGAAGAUGACAGCGAGGGUUGGAUGGAAACCAAUGAAACCUCUGCUUGAUCC .((((.....((((((......))))))....((((((((((((...---.))))))))))))...))))..((((((((((((.(....))))))....)))).)))...... ( -48.50, z-score = -4.56, R) >droGri2.scaffold_15252 15130019 111 - 17193109 GCCUCAAUAUUGGCCAGGCGAGUGGCCAUCGAGCUGUCGGAGUCGGA---GGACUCCGAUAGUGAAGAUGACAGCGAGGGUUGGAUGGAACCGAACGAAACCUCCGCUUGAUCU (((........)))((((((.((.(.((((..((((((((((((...---.))))))))))))...)))).).))((((.(((..((....))..)))..)))))))))).... ( -48.30, z-score = -3.57, R) >anoGam1.chr3R 11748001 108 + 53272125 GCCUCCAUCCUUGCCCGUCGCGUCGCCAUCGAGCUGUCCGAGUCGGA---GUCGUCCGAAAGCGACGACGACAGCGAGGGCUGGCAGGAAACGAACGAAACGUCCGCAUGA--- .(((((((((((((..((((((((((..(((..(..((((...))))---...)..)))..)))))).)))).))))))).))).)))...((.(((...))).)).....--- ( -44.40, z-score = -1.31, R) >apiMel3.Group3 1666404 114 + 12341916 GCAUCCAUAUUGGCGCGGCGAGUUGCUGUAGAAUUUAGCGACAGUGACUCAGCAUCGGAGAGCGAAUGUGACAGUGAAGGAUGGGGCGAGCAGGAAACGAAUCUCGCGUGACCC .(((((.(((((.((((((((((..((((.(.......).))))..)))).)).(((.....))).)))).)))))..)))))..(((((..(....)....)))))....... ( -39.30, z-score = -1.55, R) >triCas2.ChLG8 1963129 108 + 15773733 GCGAGUAUUUUAGCUCGGAGGGUUGCAGUCGAGUUUAGCGAUUCGGAUUCAGGAAGCGAUUCGGAGUGUGAUAGUGAGGGUUGGGGCGAA---AAUGAAACGUCGGCGUGA--- ((.......(((((((.....((..((.(((((((..((..(((.......))).)))))))))..))..)).....)))))))((((..---.......)))).))....--- ( -24.80, z-score = 1.00, R) >consensus GCAUCAAUCUUGGCCAGACGUGUGGCUAUUGAGUUAUCCGAGUCGGA___GGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACAUCGGCUUGAUCC ((..........((..........))..(((..(((((((((((.......))))))))))).........((((....))))............))).......))....... (-17.35 = -17.17 + -0.18)
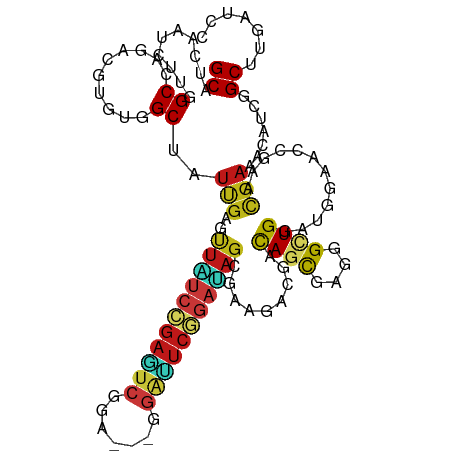
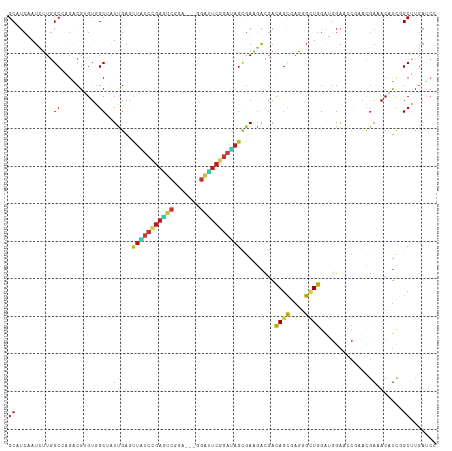
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:22 2011