| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,929,559 – 10,929,625 |
| Length | 66 |
| Max. P | 0.904942 |

| Location | 10,929,559 – 10,929,625 |
|---|---|
| Length | 66 |
| Sequences | 9 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Shannon entropy | 0.37871 |
| G+C content | 0.28620 |
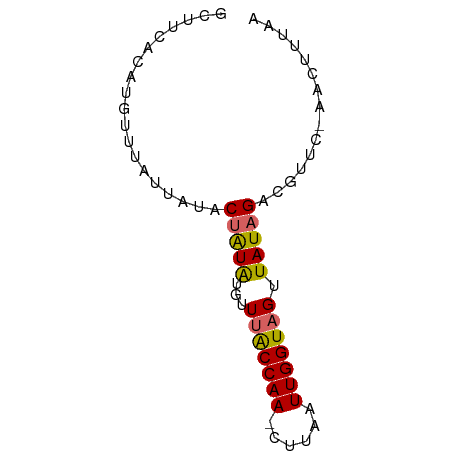
| Mean single sequence MFE | -11.46 |
| Consensus MFE | -8.23 |
| Energy contribution | -8.38 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
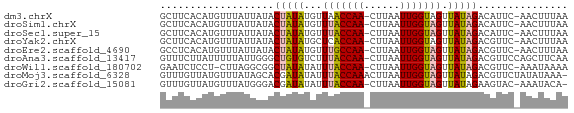
>dm3.chrX 10929559 66 + 22422827 GCUUCACAUGUUUAUUAUACUAUAUGUUAACCAA-CUUAAUUGGUAGUUAUAGACAUUC-AACUUUAA .......((((((((...(((((..(((((....-.)))))..))))).))))))))..-........ ( -10.30, z-score = -1.72, R) >droSim1.chrX 8482623 66 + 17042790 GCUUCACAUGUUUAUUAUACUAUAUGUUUACCAA-CUUAAUUGGUAGUUAUAGACAUUC-AACUUUAA ...................(((((...(((((((-.....))))))).)))))......-........ ( -9.60, z-score = -1.49, R) >droSec1.super_15 1641894 66 + 1954846 GCUUCACAUGUUUAUUAUACUAUAUGUUUACCAA-CUUAAUUGGUAGUUAUAGACAUUC-AACUUUAA ...................(((((...(((((((-.....))))))).)))))......-........ ( -9.60, z-score = -1.49, R) >droYak2.chrX 10582932 66 - 21770863 GCUUCACAUGUUUAUUAUACUAUAUGCUCACCAA-CUUAAUUGGUAGUUAUAGACGUUC-AACUUUAA ...................(((((.(((.(((((-.....)))))))))))))......-........ ( -9.30, z-score = -1.24, R) >droEre2.scaffold_4690 14553997 66 - 18748788 GCCUCACAUGUUUAUUAUACUAUAUGUUUGCCAA-CUUAAUUGGUAGUUAUAGACGUUC-AACUUUAA ...................(((((...(((((((-.....))))))).)))))......-........ ( -9.60, z-score = -0.83, R) >droAna3.scaffold_13417 5972792 67 - 6960332 GUUUCUUAUUUUUAUUGGGCUGUGUCUUUACCAA-CUUAAUUGGUAGUUAUAGACGUUCCAGCUUCAA ..............((((((((((((((((((((-.....)))))))....)))))...)))).)))) ( -14.90, z-score = -2.56, R) >droWil1.scaffold_180702 988579 65 + 4511350 GAAUCUCCU-CUUAGGCGGCUAUAUAUUUACCAA-CUUAAUUGGUAGUUAUAGACGUUC-AAAUAAAA .........-.((..(((.(((((...(((((((-.....))))))).))))).)))..-))...... ( -13.80, z-score = -2.41, R) >droMoj3.scaffold_6328 824293 67 - 4453435 GUUUGUUAUGUUUAUAGCACGAUAUAUUUACCAAACUUAAUUGGUAGUUAUAGACGUUCUAUAUAAA- ............(((((.(((.((((.(((((((......))))))).))))..))).)))))....- ( -15.10, z-score = -2.77, R) >droGri2.scaffold_15081 3068029 65 + 4274704 GUUUGUUAUGUUUAUGGGACGAUAUAUUUACCAA-CUUAAUUGGUAGUUAUAGAAGUAC-AAAUACA- ((((((..((((.....)))).((((.(((((((-.....))))))).)))).....))-))))...- ( -10.90, z-score = -0.64, R) >consensus GCUUCACAUGUUUAUUAUACUAUAUGUUUACCAA_CUUAAUUGGUAGUUAUAGACGUUC_AACUUUAA ...................(((((...(((((((......))))))).)))))............... ( -8.23 = -8.38 + 0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:59 2011