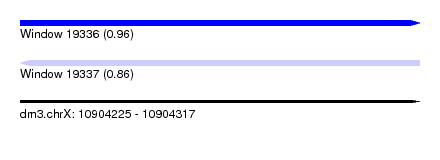
| Sequence ID | dm3.chrX |
|---|---|
| Location | 10,904,225 – 10,904,317 |
| Length | 92 |
| Max. P | 0.958887 |

| Location | 10,904,225 – 10,904,317 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.67786 |
| G+C content | 0.50013 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -11.13 |
| Energy contribution | -10.71 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
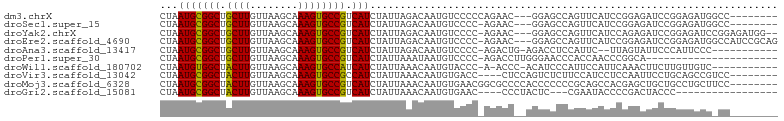
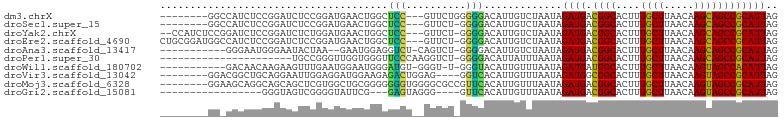
>dm3.chrX 10904225 92 + 22422827 CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAGACAAUGUCCCCCAGAAC---GGAGCCAGUUCAUCCGGAGAUCCGGAGAUGGCC-------- ((((((((((.((((........))))))))).....)))))......(((........---)))((((.....(((((....)))))..)))).-------- ( -28.30, z-score = -1.47, R) >droSec1.super_15 1615867 91 + 1954846 CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAGACAAUGUCCCC-AGAAC---GGAGCCAGUUCAUCCGGAGAUCCGGAGAUGGCC-------- ((((((((((.((((........))))))))).....)))))......(((..-.....---)))((((.....(((((....)))))..)))).-------- ( -28.00, z-score = -1.40, R) >droYak2.chrX 10556604 97 - 21770863 CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAGACAAUGUCCCC-AGAAC---GGAGCCAGUUCAUCCAGAGAUCCGGAGAUCCGGAGAUGG-- ((((((((((.((((........))))))))).....)))))...........-.((((---.......))))..(((....(((((....)))))..)))-- ( -26.90, z-score = -0.78, R) >droEre2.scaffold_4690 14528083 99 - 18748788 CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAGACAAUGUCCCC-AGAAC---GGAGCCAGUUCAUCCGGAGAUCCGGAGAUGGCCAUCCGCAG ....(((((.((((.....))))..(.((((((....................-.((((---.......)))).(((((....))))))))))))..))))). ( -33.20, z-score = -2.33, R) >droAna3.scaffold_13417 5932909 88 - 6960332 CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAGACAAUGUCCCC-AGACUG-AGACCUCCAUUC--UUAGUAUUCCCAUUCCC----------- ((((((((((.((((........))))))))).....)))))..((((.....-..((((-(((.......))--))))).....))))...----------- ( -18.10, z-score = -1.80, R) >droPer1.super_30 487236 80 - 958394 CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAAAUAAUGUCCCC-AGACCUUGGGAACCCACCAACCCGGCA---------------------- ...(((((((.((((........)))))))).)))............((.(((-((...))))).))...((.....))..---------------------- ( -15.80, z-score = 0.50, R) >droWil1.scaffold_180702 956598 89 + 4511350 CUAAUGUGGCUACUUGUUAAGCAAAGUGCCAUCAUCUAUUAAACAAUGUACCC-A-ACCC-ACAUCCCAUUCCAUUCAAACUUCUUGUUGUC----------- ...(((((((.((((........)))))))).)))..................-.-....-...............................----------- ( -9.60, z-score = -0.14, R) >droVir3.scaffold_13042 1959242 91 - 5191987 CUAAUGCGGCUACUUGUUAAGCAAAGUGCCGCCAUCUAUUAAACAAUGUGACC----CUCCAGUCUCUUCCAUCCUCCAAUUCCUGCAGCCGUCC-------- .....(((((.((((........))))))))).................(((.----((.(((....................))).))..))).-------- ( -14.45, z-score = -1.11, R) >droMoj3.scaffold_6328 783293 95 - 4453435 CUAAUGCGGCUACUUGUUAAGCAAAGUGCCGUCAUCUAUUAAACAAUGUGAACGGCGCCCCACCCCCCCGCAGCCACGAGCUGCUGCCUGCUUCC-------- .....(((((...............((((((((((............))).)))))))...........(((((.....))))).))).))....-------- ( -26.00, z-score = -1.62, R) >droGri2.scaffold_15081 3037982 79 + 4274704 CUAAUGCGGCUACUUGUUAAGCAAAGUGCCGUCAUCUAUUAAACAAUGUGAAC----CCCUACUC---CGAAUACCCCGACUACCC----------------- ...(((((((.((((........)))))))).)))..................----........---..................----------------- ( -11.10, z-score = -1.09, R) >consensus CUAAUGCGGCUGCUUGUUAAGCAAAGUGCCGUCAUCUAUUAAACAAUGUCCCC_AGACC__AGGAGCCAGUUCAUCCGGAGUUCCGGAGAUG_CC________ ...(((((((.((((........)))))))).))).................................................................... (-11.13 = -10.71 + -0.42)
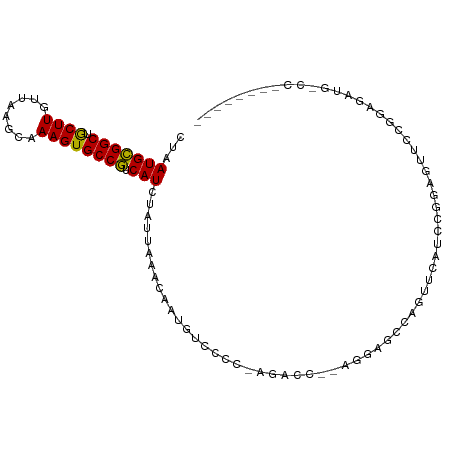
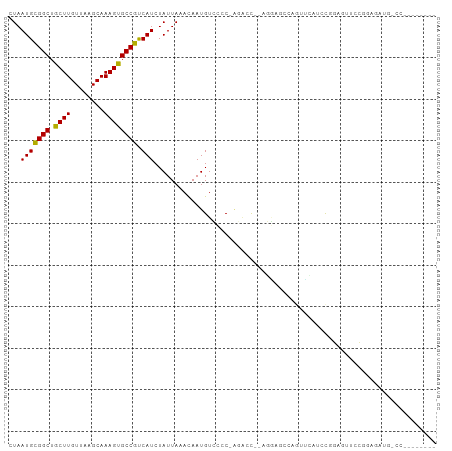
| Location | 10,904,225 – 10,904,317 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.67786 |
| G+C content | 0.50013 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.18 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
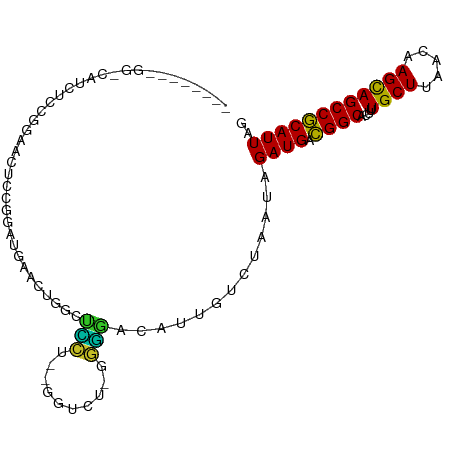
>dm3.chrX 10904225 92 - 22422827 --------GGCCAUCUCCGGAUCUCCGGAUGAACUGGCUCC---GUUCUGGGGGACAUUGUCUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG --------((((((((((((....))))).))..)))))..---..((((..((((...))))..))))((.((((....((((.....)))))))))).... ( -31.00, z-score = -1.85, R) >droSec1.super_15 1615867 91 - 1954846 --------GGCCAUCUCCGGAUCUCCGGAUGAACUGGCUCC---GUUCU-GGGGACAUUGUCUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG --------((((((((((((....))))).))..)))))..---....(-(....))..........((((.((((....((((.....)))))))))))).. ( -29.30, z-score = -1.65, R) >droYak2.chrX 10556604 97 + 21770863 --CCAUCUCCGGAUCUCCGGAUCUCUGGAUGAACUGGCUCC---GUUCU-GGGGACAUUGUCUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG --...(((((((((((((((....))))).))...(....)---..)))-)))))............((((.((((....((((.....)))))))))))).. ( -29.10, z-score = -1.26, R) >droEre2.scaffold_4690 14528083 99 + 18748788 CUGCGGAUGGCCAUCUCCGGAUCUCCGGAUGAACUGGCUCC---GUUCU-GGGGACAUUGUCUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG ..(((((..(((((((((((....))))).))..)))))))---))..(-(....))..........((((.((((....((((.....)))))))))))).. ( -34.70, z-score = -2.13, R) >droAna3.scaffold_13417 5932909 88 + 6960332 -----------GGGAAUGGGAAUACUAA--GAAUGGAGGUCU-CAGUCU-GGGGACAUUGUCUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG -----------.(((.(((((...(((.--...)))...)))-)).)))-..((((...))))....((((.((((....((((.....)))))))))))).. ( -21.20, z-score = -0.66, R) >droPer1.super_30 487236 80 + 958394 ----------------------UGCCGGGUUGGUGGGUUCCCAAGGUCU-GGGGACAUUAUUUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG ----------------------.(((.....)))..(((((((.....)-))))))...........((((.((((....((((.....)))))))))))).. ( -23.90, z-score = -0.85, R) >droWil1.scaffold_180702 956598 89 - 4511350 -----------GACAACAAGAAGUUUGAAUGGAAUGGGAUGU-GGGU-U-GGGUACAUUGUUUAAUAGAUGAUGGCACUUUGCUUAACAAGUAGCCACAUUAG -----------..........................(((((-((((-(-((((((((..((.....))..)))......))))))))......))))))).. ( -15.60, z-score = 0.37, R) >droVir3.scaffold_13042 1959242 91 + 5191987 --------GGACGGCUGCAGGAAUUGGAGGAUGGAAGAGACUGGAG----GGUCACAUUGUUUAAUAGAUGGCGGCACUUUGCUUAACAAGUAGCCGCAUUAG --------.(.(((((((....((((((.((((.....((((....----)))).)))).))))))...((..(((.....)))...)).))))))))..... ( -24.50, z-score = -1.76, R) >droMoj3.scaffold_6328 783293 95 + 4453435 --------GGAAGCAGGCAGCAGCUCGUGGCUGCGGGGGGGUGGGGCGCCGUUCACAUUGUUUAAUAGAUGACGGCACUUUGCUUAACAAGUAGCCGCAUUAG --------....((.(((.(((((.....))))).(..(((..(((.(((((...((((........))))))))).)))..)))..).....)))))..... ( -36.40, z-score = -2.30, R) >droGri2.scaffold_15081 3037982 79 - 4274704 -----------------GGGUAGUCGGGGUAUUCG---GAGUAGGG----GUUCACAUUGUUUAAUAGAUGACGGCACUUUGCUUAACAAGUAGCCGCAUUAG -----------------.........((.((((..---((((((((----(((..((((........))))..))).))))))))....)))).))....... ( -15.60, z-score = 0.11, R) >consensus ________GG_CAUCUCCGGAACUCCGGAUGAACUGGCUCCU__GGUCU_GGGGACAUUGUCUAAUAGAUGACGGCACUUUGCUUAACAAGCAGCCGCAUUAG ..................................................(....)...........((((.((((....((((.....)))))))))))).. (-11.60 = -11.18 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:32:52 2011